Figure 2.
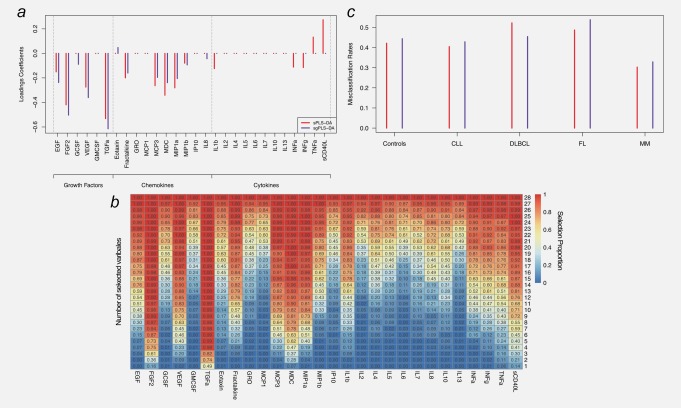
Results of the sparse and sparse group PLS‐DA (sPLS‐DA and sgPLS‐DA, respectively) analyses for all BCL cases. Loadings coefficients are presented for sPLS‐DA and sgPLS‐DA (a). Results from the stability analyses of sPLS‐DA analyses subsampling (N = 10,000 times) 80% of the study population are summarized in (b) and represents for each protein (X‐axis) and each possible number of selected variables (Y‐axis), the selection proportion (across the 10,000 repeats). Misclassification rates for each variant of the PLS algorithm used are presented in (c), for controls and each BCL subtype separately. Abbreviations: IL, interleukin; INF‐α, interferon alpha; INF‐γ, interferon gamma; GMCSF, granulocyte–macrophage colony stimulating factor; TNF‐α, tumor necrosis factor alpha; EGF, epidermal growth factor; FGF‐2, fibroblast growth factor 2; GCSF, granulocyte colony‐stimulating factor; GRO, melanoma growth stimulatory activity/growth‐related oncogene; IP10, INF‐γ‐induced protein 10; MCP‐1, monocyte chemotactic protein‐1; MCP‐3, monocyte chemotactic protein‐3; MDC, macrophage derived chemokine; MIP‐1α, macrophage inflammatory protein 1 alpha; MIP‐1ß, macrophage inflammatory protein 1 beta; sCD40L, soluble CD40 ligand; VEGF, vascular endothelial growth factor; TGF‐α, transforming growth factor alpha. [Color figure can be viewed at http://wileyonlinelibrary.com]
