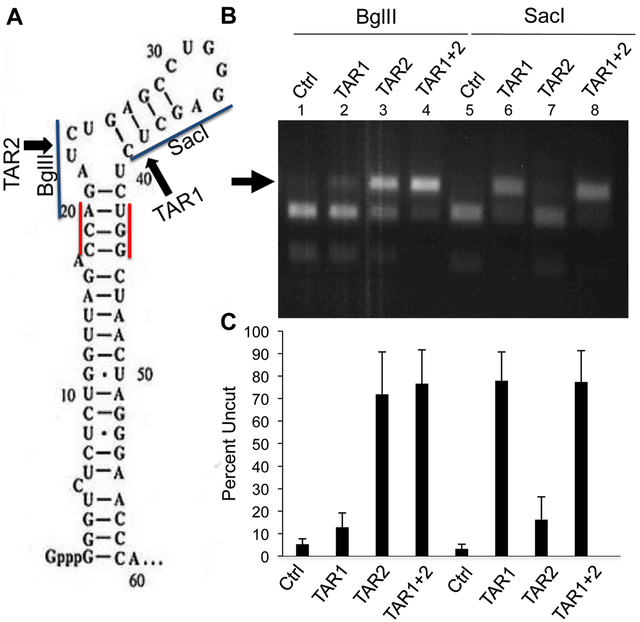
Fig. 2: Efficient cleavage of DNA encoding the HIV-1 TAR element.
(A) Schematic of the HIV-1 TAR RNA structure with the two Sp Cas9 PAM sequences, the predicted Sp Cas9 cleavage sites and relevant restriction sites indicated (B) The DNA region encoding TAR was recovered by PCR from 293T cells infected with the NL-NLuc-HXB indicator virus, as described in Fig. 1D, that also expressed Sp Cas9 and one or both sgRNAs specific for TAR, as indicated in panel A. DNA editing efficiency was then assessed by cleavage with either BglII or SacI, which cleave at sites that, in wild type TAR, underlie the predicted target sites for the TAR1 (SacI) or TAR2 (BglII) sgRNAs, as indicated in panel A. Therefore, Cas9 cleavage, if followed by the introduction of an indel during NHEJ-mediated repair, would preclude cleavage by SacI and/or BglII. A representative experiment is shown with the percentage of uncut TAR DNA indicated below. (C) Same as panel B except that this bar graph shows the average level of uncut TAR DNA, after incubation with SacI or BglII, from three independent biological replicates with SD indicated.