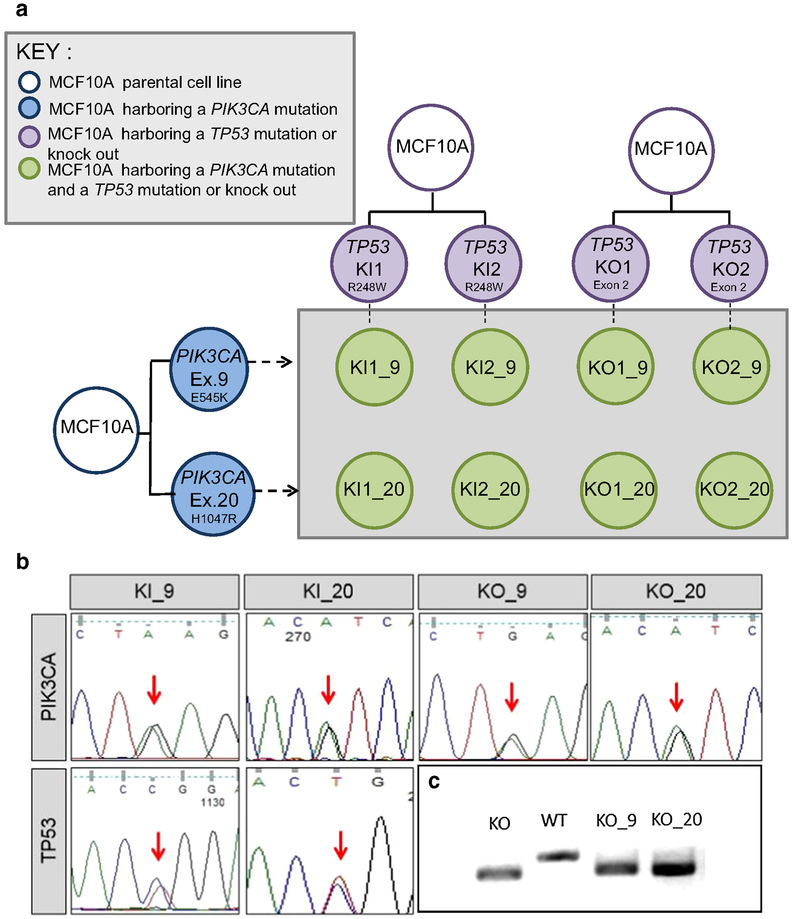
Fig. 1.
Derivation and confirmation of cell lines used in this study. a A panel of isogenic cell lines was developed with both a PIK3CA and TP53 alteration. Single TP53-altered cell lines shown in purple contain either a homozygous deletion/knockout (KO) or a TP53 R248W hotspot mutation/knockin (KI). TP53-altered cell lines were used as the parental cell line and were infected with AAV vectors containing either the PIK3CA E545K mutation in exon 9 (Ex. 9) or the H1047R mutation in exon 20 (Ex. 20) (blue). Double-mutant cell lines were developed (green) from independently derived parental clones. Nomenclature for the derived cell lines indicates the TP53 status and clone number followed by an underscore and the designated PIK3CA mutation. b Representative Sanger sequencing traces confirming mutations. Traces for the PIK3CA and TP53 mutation status for representative double-mutant clones are shown. Equal peaks (indicated by the red arrows) indicate 50:50 allelic ratios demonstrating that the mutation incorporated into one of the two alleles. c TP53 homozygous deletion is detected through deletion of a small portion of genomic DNA in exon 2 as detected by PCR and gel electrophoresis. The lower bands indicate a smaller DNA fragment due to the loss of the deleted DNA. KO = TP53 knockout, WT = TP53 wild-type parental control (MCF10A), KO_9 = TP53 knockout with PIK3CA E545K mutation, KO_20 = TP53 knockout with PIK3CA H1047R mutation