| Data collection statisticsa | thaumatin (Frame 1-20) | glucose isomerase (Frame 1-100) | thioredoxin (Frame 1-10) |
| Beamline | P14 | ||
| Wavelength [Å] | 0.96863 | ||
| Space group | P41212 | I222 | P42212 |
| Unit cell parameters: a = b, c [Å] | 58.62, 151.48 | 93.91, 99.60, 103.04 | 58.45, 151.59 |
| Number of crystals | 101 | 41 | 34 |
| Total oscillation [°] | 10 | 10 | 10 |
| Resolution [Å] | 30.1.1989 (1.95 – 1.89) | 30.1.1975 (1.80 – 1.75) | 30.3.2000 (3.20 – 3.00) |
| Temperature [K] | 296 | 296 | 296 |
| R p.i.m.b | 7.5 (25.5) | 8.8 (28.0) | 9.1 (33.2) |
| Measured reflections | 1553200 | 690000 | 1111196 |
| Unique reflections | 21850 | 48942 | 44449 |
| Average I/σ(I) | 6.07 (1.78) | 5.85 (1.66) | 4.08 (1.47) |
| Mn(I) half-set correlation CC(1/2) | 96.2 (72.2) | 95.8 (68.2) | 97.9 (75.3) |
| Completeness [%] | 99.8 (100.0) | 100.0 (99.9) | 99.9 (100.0) |
| Redundancy | 71.1 | 14.1 | 25 |
| Refinement statistics | |||
| Resolution range [Å] | 1/30/1989 | 1/30/1975 | 3/30/2000 |
| R/ Rfree [%] | 18.8/23.9 | 18.1/20.5 | 18.9/23.1 |
| Protein atoms | 1550 | 3045 | 1129 |
| Water molecules | 51 | 111 | 164 |
| Ligand molecules | 20 | 0 | 0 |
| Rms deviation | |||
| Bond-length [Å] | 0.02 | 0.026 | 0.01 |
| Bond angle [°] | 2.04 | 2.22 | 1.43 |
| B factor [Å2] | |||
| Protein | 22.6 | 20 | 50 |
| Water | 25.1 | 27.1 | 29.7 |
| Ligand | 20.4 | ||
| Ramachandran plot analysis | |||
| Most favored regions [%] | 97.67 | 95.32 | 96.13 |
| Allowed regions [%] | 2.44 | 4.16 | 3.64 |
| Generously allowed regions [%] | 0.49 | 0.52 | 0.23 |
| a: Values in parentheses are for the highest resolution shell. | |||
b: (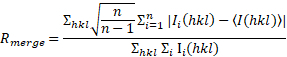 ), where I (hkl) is the mean intensity of the reflections hkl, Σhkl is the sum over all reflections and Σi is the sum over i measurements of reflection hkl. ), where I (hkl) is the mean intensity of the reflections hkl, Σhkl is the sum over all reflections and Σi is the sum over i measurements of reflection hkl. |

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.