Figure 1.
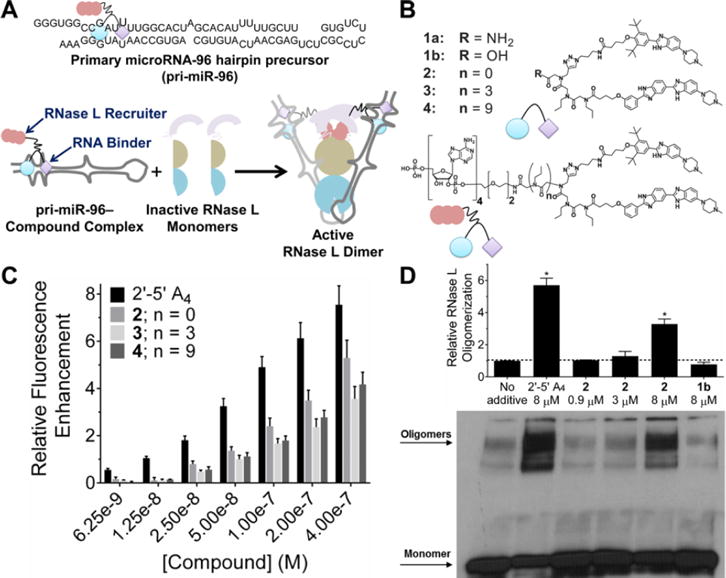
Design and characterization of a transcript-selective RNase L recruiting compound. (A) Top, secondary structure of the primary transcript of microRNA-96 (pri-miR-96). Bottom, schematic depiction of active RNase L recruitment to pri-miR-96 by 2. (B) Structures of compounds used in this study. (C) In vitro cleavage of pri-miR-96 by 2′-5′A4 and chimeric small molecules with different linker lengths; n = 0 linker (2) displayed the highest cleavage capability. (D) Representative Western blot and quantification of cross-linked monomer and oligomer (active) forms of RNase L upon treatment with 2′-5′A4, 2 (selective for pri-miR-96), or lb which lacks 2′-5′A4. Data are expressed as mean ± s.e.m. (n > 3). *p < 0.05, as measured by a two-tailed Student t test.
