Figure 2.
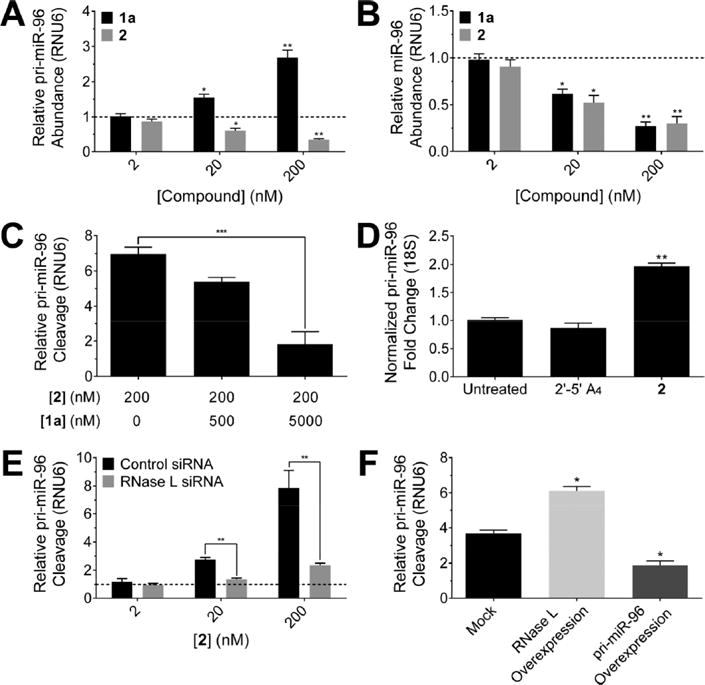
Small molecule RNase L recruitment shows on-target effects in cells. (A) Treatment of MDA-MB-231 triple negative breast cancer cells with 2 decreased abundance of pri-miR-96 via cleavage, as measured by RT-qPCR In contrast, la boosted levels of pri-miR-96 by inhibiting Drosha processing (lacks 2,-5′A4). (B) Effect of la and 2 on mature miR-96 levels. (C) 2-mediated cleavage of pri-miR-96 is reduced by addition of la. The effect of la itself was accounted for in the calculation. (D) RT-qPCR of RNAs isolated from immunopreci- pitated RNase L protein after cells were treated with 2′-5′A4 or 2 (200 nM). RNAs bound to RNase L treated with 2 show enrichment of the pri-miR-96 transcript normalized to RNA immunoprecipitated from ¡5- actin. (E) Relative cleavage of pri-miR-96 by 2 upon knock down of RNase L by siRNA (F) RNase L overexpression resulted in increased cleavage activity of 2 (20 nM), whereas overexpression of pri-miR-96 resulted in decreased cleavage activity. Data are expressed as mean ± s.e.m. (n > 3). *p < 0.05, **p < 0.01, as measured by a two-tailed Student t test.
