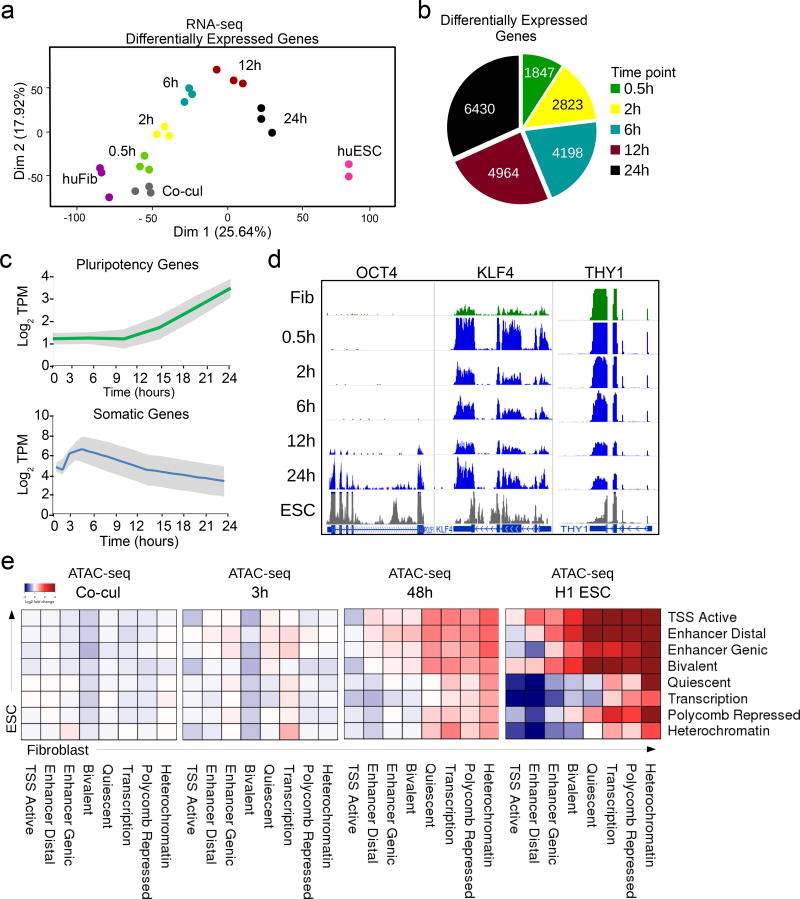
Figure 1. Nuclear reprogramming of human fibroblasts after fusion with mouse ESCs.
(a) Principal component analysis plot of RNA-seq data shows a continuous trajectory of transcriptome changes in human fibroblasts towards human ESCs over time post-fusion in heterokaryons (n= 3 biological replicates). (b) Pie chart depicting the number of differentially expressed (DE) human genes at each time point during early reprogramming. (c) Line plot showing average RNA expression during reprogramming from 331 somatic and 597 embryonic associated genes taken from the molecular signature database (MSigDB). Gray shades represent the standard deviation from the median. (d) RNA-seq tracks for human OCT4, KLF4, and THY1 genes over the time course of heterokaryon reprogramming. (n= 3 biological replicates). (e) Chromatin state transition matrix. Using chromatin states mapped and defined by the Roadmap Epigenomics Consortium, the heatmap represents the transition from human fibroblast (columns) to embryonic stem cell (rows). The color spectrum from blue to red represents the log-fold change of the median ATAC-seq peak signal of each transition at each time point relative to time 0 (fibroblast alone).