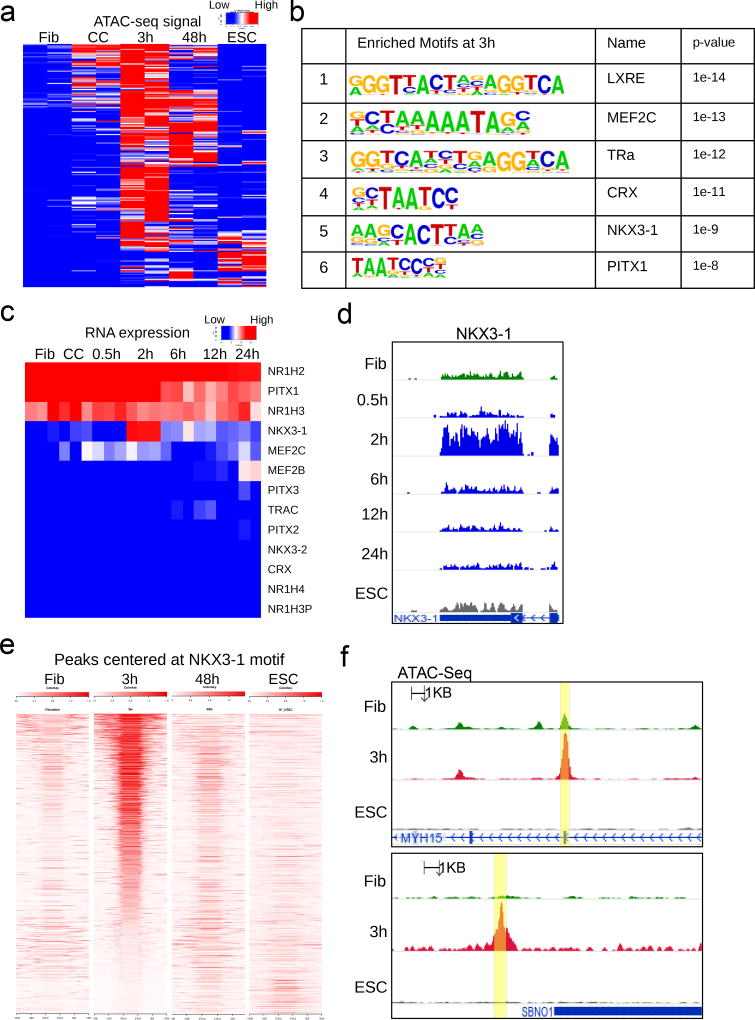
Figure 2. Motifs at early accessible chromatin and gene expression dynamics identify NKX3-1.
(a) Heatmap of ATAC-seq signal at genomic regions that become more accessible at 3h post-fusion heterokaryons (mouse ESC × human fibroblast) compared to unfused human fibroblast. Signals from those same regions are also shown for human fibroblast, human fibroblast from mouse ESC co-culture, 48h post-fusion, and human ESC as a reference. (b) Motif enrichment at 3h peaks that increased in accessibility, using fibroblast peaks as background. Motif enrichment was calculated with the cumulative binomial distribution at 3h peaks that increased in accessibility, using fibroblast as background. P-value is not adjusted for multiple testing. Benjamini-adjusted q-value calculated by Homer software is reported as 0.0000 for all motifs shown. (n = 53043 target sequences) (c) Expression by RNA-seq of candidate genes obtained from motif enrichment in 2b and their close family members. (d) RNA-seq signal track at the human NKX3-1 locus over the time course of heterokaryon reprogramming. (n= 3 biological replicates). (e) Density heatmap of ATAC-seq signal at 1705 genomic regions, centered at the NKX3-1 motif and ranked by accessibility at three hours. (f) ATAC-seq tracks of two representative peaks containing the NKX3-1 motif shown in 2e (yellow highlights). (n= 3 biological replicates).