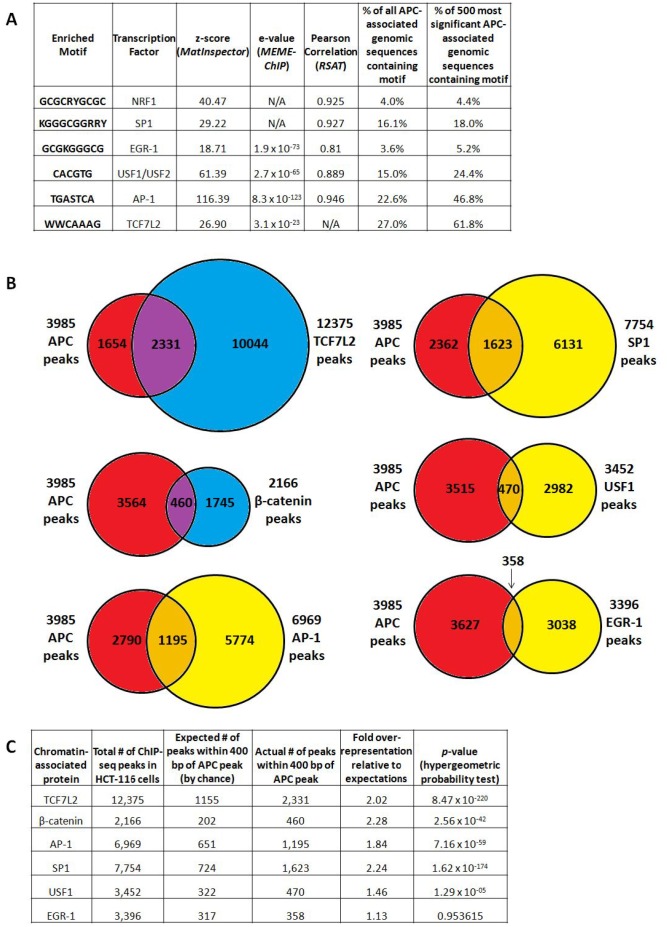
Figure 3. APC-associated genomic sequences are enriched for TCF7L2, AP-1 and SP1 transcription factor binding motifs.
(A) Motif analysis of all peaks using the MatInspector (Genomatix Software Suite), MEME-ChIP and Regulatory Sequence Analysis Tools (RSAT) algorithms detected significant enrichment of predicted binding sites, particularly for the AP-1, EGR-1 and USF1/USF2 transcription factors. All transcription factor binding motifs occurred more frequently in the subset of 500 genomic sequences enriched by APC ChIP with the lowest p-values, particularly those for TCF7L2 (as expected) and AP-1. (B) 3,985 APC ChIP-seq peaks were compared with corresponding peaks from several transcription factor ChIP-seq experiments. (C) Overlap data were tabulated to calculate fold overrepresentation of each transcription factor binding site among APC-associated genomic regions, relative to expected background (p-values calculated by hypergeometric probability test).