Figure 2.
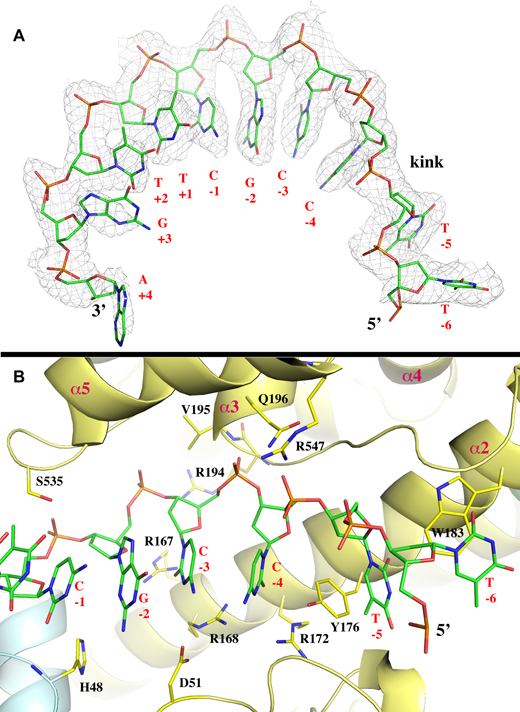
Conformation of ssDNA substrate and its interaction with MtTOP1-704t. (A) The ssDNA bound to MtbTOP1-704t resembles a B-DNA conformation, which is interrupted between –4 and –5 positions. The 2Fo-Fc electron density map is drawn in gray mesh and contoured at 1σ level and within 2.1 Å of the ssDNA, which is drawn in stick format. (B) Interaction between ssDNA and MtbTOP1-704t including the creation of a ssDNA kink and the C nucleotide binding pocket at –4 position. Several arginine residues involved in the interaction with either the phosphate backbone (R194 and R547) or bases (R167, R168 and R172) are presented. One anchoring site of ssDNA to MtTOP1-704t is located at the phosphate group between –3 and –4 positions. The phosphate group is positioned below the N-terminus of the α3 helix of D4, which has an equivalent charge of about +0.5 unit. In the apo structures of MtbTOP1-704t, a phosphate group or a sulfide group from crystallization buffer is commonly found at this position. The tyrosine Y176 inserts between the bases at –4 and –5 positions, helping create a kink in the base packing of ssDNA. The residue Y176 forms π–π stacking with bases at both –4 and –5 positions. R168 and R172 provide specific interactions to the C nucleotide at –4 position. Both Figure 2A and B were prepared based on the MtTOP1-704t/MTS2–11 structure (PDB code: 6CQI, Table 1).
