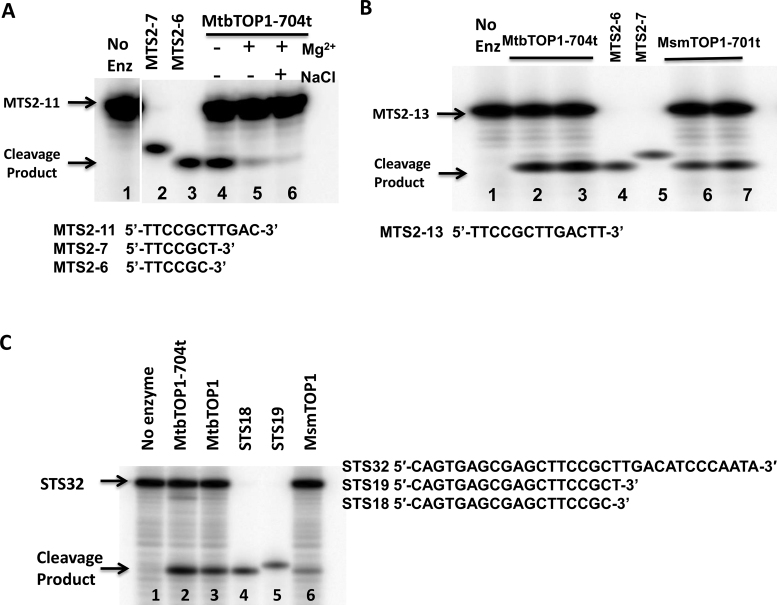
Figure 5.
Mapping of exact cleavage site on single-stranded oligonucleotide substrates. (A) Cleavage and religation of MTS2–11 substrate by MtbTOP1-704t. Lane 1: MTS2–11 substrate with no enzyme added. Lane 2: MTS2-7 oligo standard. Lane 3: MTS2-6 oligo standard. Lane 4: cleavage by 1 pmol MtbTOP1-704t. Lane 5: cleavage by 1pmol MtbTOP1-704t with 0.5 mM Mg2+ added for DNA religation. Lane 6: cleavage by 1pmol MtbTOP1-704t followed by addition of 0.5 mM Mg2+ and 1 M NaCl to dissociate the enzyme following DNA religation. (B) Cleavage of MTS2-13 by MtbTOP1-704t and MsmTOP1-701t. Lane 1: MTS2-13 substrate with no enzyme added. Lanes 2, 3: 1 and 2 pmol of MtbTOP1-704t added. Lanes 4: MTS2-6 oligo standard. Lane 5: MTS2-7 oligo standard. Lanes 6, 7: 1 and 2 pmol of MsmTOP1-701t added. (C) Cleavage product with STS32 oligonucleotide substrate. Lane 1: STS32 substrate with no enzyme added. Lanes 2, 3 and 6: 1 pmol MtbTOP1-704t, MtbTOP1 and MsmTOP1 added respectively. Lanes 4: STS18 oligo standard. Lane 5: STS19 oligo standard.