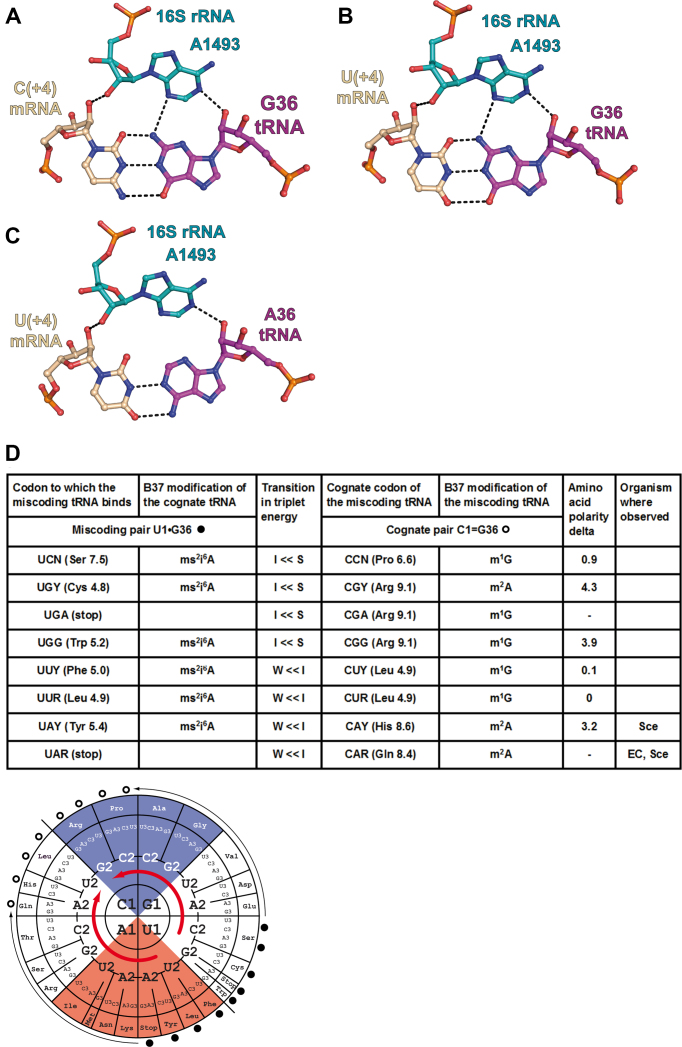
Figure 3.
Substitution of a C1 = G36 pair or a U1-A36 pair by a U1•G36 base pair in the first position of the codon–anticodon duplex. (A) Cognate C1 = G36 pair from the complex of 70S ribosome with mRNA and tRNALeu (PDB ID: 4v87); (B) near-cognate U1•G36 pair from the complex of 70S ribosome with mRNA and tRNALeu (PDB ID: 4v8b); (C) cognate U1-A36 pair from the complex of 70S ribosome with mRNA and tRNATyr (PDB ID: 4v8d). Same comments as for Figure 2. (D, top) The in vivo data for Escherichia coli and Saccharomyces cerevisiae are from (6,7) and (28); those for the CHO cells are from (27). (D, bottom) The miscoding pairs (dark circles) are indicated with the thin arrows pointing to the miscoding tRNA (open circles) (e.g. a tRNAHis miscodes a Tyr codon). The thick red arrows indicate the energetic transitions between Intermediate (white) and Strong (blue) codon–anticodon triplets as well as between Weak (red) and Intermediate ones. Because the substitutions are at the first position, the transitions start in the bottom right quadrant and end in the top left quadrant. The miscoding tRNAs are good binders and replace weaker, but more modified, tRNA binders. Only two instances could be found in the literature.