Abstract
Trans-differentiation of one somatic cell type into another has enormous potential to model and treat human diseases. Previous studies have shown that mouse embryonic, dermal, and cardiac fibroblasts can be reprogrammed into functional induced-cardiomyocyte-like cells (iCMs) through overexpression of cardiogenic transcription factors including GATA4, Hand2, Mef2c, and Tbx5 both in vitro and in vivo. However, these previous studies have shown relatively low efficiency. In order to restore heart function following injury, mechanisms governing cardiac reprogramming must be elucidated to increase efficiency and maturation of iCMs.
We previously demonstrated that inhibition of pro-fibrotic signaling dramatically increases reprogramming efficiency. Here, we detail methods to achieve a reprogramming efficiency of up to 60%. Furthermore, we describe several methods including flow cytometry, immunofluorescent imaging, and calcium imaging to quantify reprogramming efficiency and maturation of reprogrammed fibroblasts. Using the protocol detailed here, mechanistic studies can be undertaken to determine positive and negative regulators of cardiac reprogramming. These studies may identify signaling pathways that can be targeted to promote reprogramming efficiency and maturation, which could lead to novel cell therapies to treat human heart disease.
Keywords: This Month in JoVE, Issue 136, Cardiac reprogramming, transcription factors, microRNAs, pro-fibrotic signaling, compound, TGF-β receptor 1 inhibitor
Introduction
Ischemic heart disease is a leading cause of death in the United States1. Approximately 800,000 Americans experience a first or recurrent myocardial infarction (MI) per year1. Following MI, the death of cardiomyocytes (CMs) and cardiac fibrosis, deposited by activated cardiac fibroblasts, impair heart function2,3. Progression of heart failure following MI is largely irreversible due to the poor regenerative capacity of adult CMs4,5. While current clinical therapies slow disease progression and decrease risk of future cardiac events6,7,8,9, no therapies reverse disease progression due to the inability to regenerate CMs post-infarction10. Novel cell therapies are emerging to treat patients following MI. Disappointingly, clinical trials delivering stem cells to the heart following MI thus far have shown inconclusive regenerative potential11,12,13,14,15,16,17,18.
The generation of human-derived induced pluripotent stem cells (hiPSCs) from fibroblasts by overexpression of four transcription factors, first demonstrated by Takahashi & Yamanaka, opened the door to new breakthroughs in cell therapy19. These cells can differentiate into all three germ layers19, and several highly efficient methods for generating large numbers of CMs have been previously shown20,21. HiPSC-derived CMs (hiPS-CMs) offers a powerful platform to study cardiomyogenesis and may have important implications for repairing the heart following injury. However, hiPS-CMs currently face translational hurdles due to concerns of teratoma formation22, and their immature nature may be pro-arrhythmogenic23. Reprogramming fibroblasts into hiPSCs sparked interest in directly reprogramming fibroblasts into other cell types. Ieda et al. demonstrated that overexpression of GATA4, Mef2c, and Tbx5 (GMT) in fibroblasts results in direct reprogramming to cardiac lineage, albeit at low efficiency24. Reprogramming efficiency was improved with the addition of Hand2 (GHMT)25. Since these early studies, many publications have demonstrated that altering the reprogramming factor cocktail with additional transcription factors26,27,28,29, chromatin modifiers30,31, microRNAs32,33, or small molecules34 leads to improved reprogramming efficiency and/or maturation of induced cardiomyocyte-like cells (iCMs).
Here we provide a detailed protocol to generate iCMs from mouse embryonic fibroblasts (MEFs) with high efficiency. We previously showed that the GHMT cocktail is significantly improved with the addition of miR-1 and miR-133 (GHMT2m) and is further improved when pro-fibrotic signaling pathways including transforming growth factor β (TGF-β) signaling or Rho-associated protein kinase (ROCK) signaling pathways are inhibited35. Using this protocol, we show that approximately 60% of cells express cardiac Troponin T (cTnT), approximately 50% express α-actinin, and a high number of beating cells can be observed as early as Day 11 following transduction of reprogramming factors and treatment with the TGF-β type I receptor inhibitor A-83-01. Furthermore, these iCMs express gap junction proteins including connexin 43 and exhibit spontaneous contraction and calcium transients. This marked improvement in reprogramming efficiency compared to earlier studies demonstrates the potential to regenerate CMs from endogenous cell populations that remain in the heart post-infarction.
Protocol
All experiments requiring animals were approved by the Institutional Animal Care and Use Committee at the UC Denver Anschutz Medical Campus.
1. Isolation of MEFs
Purchase C57BL/6 pregnant mice at E13. Ship overnight.
Euthanize the mother according to approved IACUC protocols (ex: ~1.3 L/min CO2 until animal appears dead followed by cervical dislocation)
Spray the mother with 70% ethanol and open abdominal cavity. Remove the uterine horn containing embryos and place in a 10 cm dish with sterile PBS.
Make an incision in the embryo sac to release embryo. Transfer embryo to clean 10 cm dish with sterile PBS. Rinse thoroughly.
Cut the body below the liver and discard the head and upper body. Remove internal organs and discard. Transfer the remaining tissue to a clean 10 cm dish with sterile PBS and transfer the plate to a Biosafety Level 1 or 2 Cabinet. Combine tissues from all embryos in a clean, dry 10 cm dish and mince into fine pieces.
Add 6 mL of 0.25% trypsin/1 mM EDTA to minced embryos. Triturate several times by pipette to break up tissue. Incubate for 40 min at 37 ˚C with 5% CO2.
Resuspend cells in growth medium (Table 1). The volume of media is adjusted based on the number of embryos harvested. In general, use a ratio of approximately 25 mL growth media per 1.2 embryos.
Plate 25 mL of resuspended cells in a 15 cm dish. After 24 h, aspirate the media and add 25 mL of fresh growth medium to each plate.
After 72 h, add 2 mL of 0.25% trypsin/1 mM EDTA to each 15 cm dish. When cells detach from the culture dish, inactivate trypsin with 15 mL growth medium and collect cells in 50 mL polystyrene conical tubes. Centrifuge cells for 3 min at 160 x g at room temperature. Resuspend the cell pellet in growth medium and filter through a 70 µm pore cell strainer.
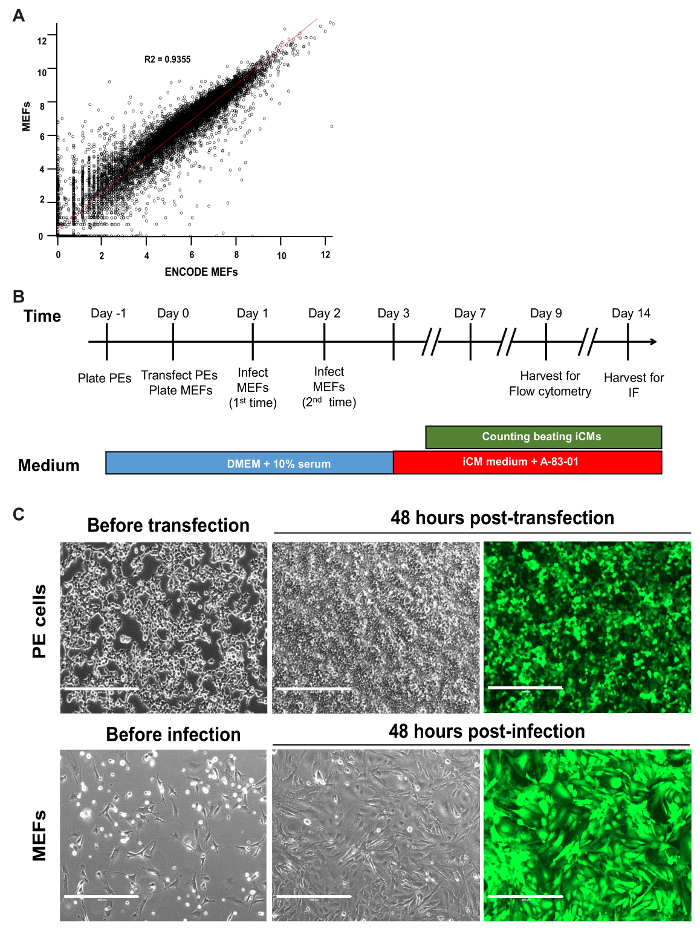
Count cells using a hemocytometer and freeze MEFs in 2 million cells aliquots in freeze medium (10% DMSO, 25% FBS in Dulbecco's Modified Eagle Medium (DMEM)/High glucose) at -80 °C. Transfer cells to liquid nitrogen after 24 h and store until ready to use. NOTE: Global gene expression in MEFs was profiled isolated using this protocol by RNA-sequencing (RNA-seq) to ensure cell authentication. RNA-seq data from ENCODE MEF cells were downloaded from NIH Gene Expression Omnibus (GEO) with accession number GSE93454. RNA-seq gene expression data for the MEFs used by us were deposited in NIH GEO with accession number GSE71405. These two data sets were merged according to common gene symbols. The expression data were then log-transformed and a scatterplot of the expression data for our MEFs and ENCODE MEFs were generated and fit with a linear regression line (Figure 1A). MEFs isolated using this protocol are molecularly similar to those isolated by other labs.
2. Production of Retrovirus and Transduction of MEFs
NOTE: All steps in this section must be carried out in a Biosafety Level 2 Cabinet. Since this protocol utilizes retroviral transduction, ensure that safety precautions are taken including treating all waste containing viral media with 10% bleach for at least 20 min. Refer to Figure 1B for a timeline for reprogramming.
- Production of Retrovirus using the Platinum E (PE) cell line
- Maintain commercially available PE cells in PE medium (Table 1) containing blasticidin and puromycin selection markers to ensure expression of gag-pol and env genes for efficient retroviral particle production36. Passage cells at 1:4 or 1:5 every two days. Take care to prevent overcrowding of cells as this can reduce retroviral production yields.
- On the day -1, seed approximately 5 x 106 cells per 10 cm dish in the growth medium (Table 1) 16-20 h prior to transfection (see Table 2 for plating densities for other standard cell culture dish sizes). Gently rock dishes back and forth several times and carefully place in a 37 °C, 5% CO2 incubator to ensure even plating distribution (see Figure 1C for optimal plating density prior to transfection). NOTE: Take care to plate PE cells in the medium free of selection markers prior to transfection to ensure medium containing generated retroviral particles does not kill MEFs.
- On the day 0, prepare DNA for transfection. For 10 cm dish transfection, add 2 µg of each reprogramming factor — GATA4, Hand2, Mef2c, Tbx5, miR-1, and miR-133 (GHMT2m) — to a 1.5 mL tube. NOTE: Refer to Table 2 to scale the amount of DNA needed to transfect other standard cell culture dish sizes.
- In a separate 1.5 mL tube, add 360 µL reduced serum media. Then add 36 µL transfection reagent directly to reduced serum media. Gently flick the tube and incubate at room temperature for 5 min. NOTE: To avoid reduced transfection efficiency, carefully add transfection reagent directly to the reduced serum media.
- Add reduced serum media/transfection reagent mixture to GHMT2m DNA cocktail. Gently flick the tube and incubate at room temperature for 15 min. Do not vortex.
- Add the reaction mixture dropwise to PE cells, gently swirl media for 10-15 s, and place in a 37 °C, 5% CO2 incubator. NOTE: According to the manufacturer, PE cells generate an average titer of 106 to 107 infectious units/mL. This protocol generates approximately 2 x 106 infections units/mL by 24 h and 6 x 106 infections units/mL by 48 h for an average MOI of 4. Transfection of GFP/DsRed may also be used as a surrogate for transfection/transduction efficiency if desired; transfection efficiency is expected to exceed 90% by 48 h (Figure 1C).
- Transduction of MEFs
- On the Day 0, coat dishes with collagen and place in a 37 °C, 5% CO2 incubator for at least 2 h.
- Remove MEFs from liquid nitrogen and thaw. Plate 0.2 x 106 cells per 60 mm dish in the growth medium. Gently rock plates to ensure even plating distribution and place in the incubator (see Figure 1C for optimal plating density and Table 2 for plating densities for other standard cell culture dish sizes). NOTE: These plating densities have been tested over multiple batches of isolated MEFs; unless MEFs display significantly compromised viability, adjusting cell seeding densities is not necessary.
- On the day 1, 24 h post-transfection, use a 30 mL syringe to collect retroviral medium generated by PE cells. Pass the medium through a 0.45 µm pore-size filter into a 50 mL conical tube. Add hexadimethrine bromide transfection reagent to a final concentration of 6 µg/mL. Carefully add 10 mL fresh growth medium to PE cells by pipetting media onto the wall of the culture dish to prevent displacement of cells.
- Aspirate the medium from MEFs and add 4 mL of freshly harvested retroviral medium to each dish. Return MEFs to the incubator. Add 10% bleach to all materials in contact with the viral medium to neutralize retroviral particles.
- On the day 2, 48 h post-transfection, repeat steps 2.2.2 and 2.2.3. PE cells are discarded following the 2nd collection of the retroviral medium. NOTE: Add 10% bleach to discarded PE cells for at least 20 min to neutralize unwanted retrovirus.
- On the day 3, aspirate medium from MEFs and replenish with 4 mL iCM medium (Table 1) containing 0.5 µM A-83-01, a TGF-β type I receptor inhibitor. Replenish iCM medium containing A-83-01 every 2 days. NOTE: If using GFP as a transduction control, the expression is expected to exceed 90% by this time point (Figure 1C).
3. Flow Cytometry for Cardiac Markers
Aspirate medium from Day 9 reprogrammed MEFs and wash (1x) with 1x PBS to remove dead cells and debris.
Add 1 mL 0.25% Trypsin/EDTA for 5 min at 37 ˚C. Check cells using a light microscope and shake dish; if cells remain attached, incubate 5 more min at 37 ˚C until cells detach.
Wash cells in 1x PBS containing 5% FBS and filter through a 70 µm pore cell strainer.
Centrifuge at 160 x g for 3 min at room temperature and aspirate liquid from the pellet. NOTE: To increase the cell yield, leave approximately 50 µL liquid above the cell pellet.
Wash cells with 500 µL 1x PBS containing 1% BSA.
Fix cells with 0.2 mL/1 million cells fixation solution for 20 min on ice.
Add 1 mL of 1x ice-cold perm/wash buffer. NOTE: The manufacturer's solution is 10x.
Centrifuge cells at 160 x g for 5 min at 4 °C and aspirate supernatant.
Repeat steps 3.7 and 3.8 an additional time. After the 2nd wash, proceed directly to next step or keep cells at 4 °C overnight.
Block with 100 µL of 5% goat serum and donkey serum in 1x perm/wash buffer for 30 min at room temperature.
Add 100 µL of diluted primary antibody. NOTE: For this protocol, cells were labeled with mouse Troponin T (1:200) and mouse α-actinin (1:200) to quantify the percentage of cells positive for key components of the cardiac sarcomere. Incubate primary antibody for 1 h at room temperature.
Centrifuge cells at 160 x g for 5 m at 4 °C, aspirate supernatant, and wash 2x with ice-cold perm/wash buffer.
Add 100 µL of diluted secondary antibody. NOTE: For this protocol, cells were labeled with anti-mouse 647 nm secondary antibody (1:200). Incubate secondary antibody for 45 min at room temperature on an end-over-end tube rotator. Protect cells from light by wrapping tubes in foil.
Centrifuge cells at 160 x g for 5 min at 4 °C, aspirate supernatant, and wash 2x with ice-cold perm/wash buffer.
Add 1 mL of 1% BSA in PBS and perform FACS immediately. NOTE: Generate a forward scatter vs side scatter plot to gate viable cells first when performing FACS. Alternatively, use a commercially available live/dead cell stain prior to fixing cells to identify live and dead cell populations.
4. Immunostaining of Reprogrammed MEFs
Aspirate medium from day 14 reprogrammed MEFs and wash (1x) with 1 mL 1x ice-cold PBS. NOTE: For immunostaining, reprogramming was performed in 12 well plates to minimize antibody solution volumes. 24 well plates may also be used; simply half all following volumes.
Aspirate and fix cells with 500 µL 2% paraformaldehyde for 10 min at room temperature.
Aspirate and wash cells 3 times with 1 mL 1x PBS (5 min per wash at room temperature).
Permeabilize cells with 500 µL 0.2% permeabilization reagent in PBS for 15 min at room temperature.
Aspirate and block in 500 µL 10% horse serum in PBS for 30 min at room temperature.
Aspirate and incubate with 500 µL of the diluted primary antibody for 1 h at room temperature. NOTE: For this protocol, cells were labeled with mouse Troponin T or α-actinin (each 1:400) and co-labeled with Connexin 43 (1:400) or Troponin I (1:400).
Aspirate primary antibody and wash three times with 1 mL 1x PBS (5 min per wash at room temperature).
Aspirate and incubate with 500 µL diluted secondary antibody for 1 hour at room temperature. NOTE: For this protocol, cells were labeled with anti-mouse 555 nm secondary antibody (1:800) to recognize Troponin T or α-actinin and anti-rabbit 488 nm secondary antibody (1:800) to recognize Connexin 43 or Troponin I. Additionally, nuclei were stained with Hoechst (1:10,000). Protect cells from light by incubating in the dark.
Aspirate the secondary antibody and wash three times with 1 mL 1x PBS (5 min per wash at room temperature). Wrap the plate in foil to protect cells from light and store at 4 °C.
Image using three-channel epifluorescence microscopy.
5. Calcium Imaging
NOTE: If imaging at 10X magnification, standard cell culture dishes and plates are suitable for calcium imaging. Imaging at higher magnification requires that cells be plated on glass coverslips or glass bottom dishes.
Aspirate medium and add 2 mL (for a 6 well plate) modified Tyrode's solution (140 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 10 mM HEPES, 0.1% BSA, and 1% pyruvate, pH 7.4) containing 5 µM Fura-2 AM and 0.1% Pluronic F-127 to beating (D14 to D20) iCMs. Incubate for 30 minutes at 37 °C. NOTE: Protect cells from light to prevent quenching of fluorescent indicator.
Wash cells in 2 mL Tyrode's solution for 30 min at room temperature to allow de-esterification of Fura-2 AM.
Image with spinning disk confocal microscopy at 340 nm and 380 nm using Slidebook 5.5 Ca2+ imaging software. Perform the imaging at room temperature.
Calcium transients are calculated using the ratio of fluorescence intensity at 340 nm over the fluorescence intensity at 380 nm.
If desired, treat iCMs with 1 or 2 µM isoproterenol or 10 µM Nifedipine to manipulate calcium handling after imaging baseline calcium transients. Add compounds locally to cells and image.
Representative Results
Using the reprogramming strategy outlined above and in Figure 1B, we generated iCMs with approximately 70% of cells expressing cardiac Troponin T and approximately 55% of cells expressing cardiac α-actinin, quantified by flow cytometry at Day 9 following transduction of GHMT2m (Figure 2A and B). Additionally, the majority of cells express cardiac Troponin T, Troponin I, and cardiac α-actinin as well as the gap junction marker Connexin 43 at Day 14 following transduction (Figure 2C and D). Imaging with higher magnification reveals well-defined sarcomere structure formation and gap junctions forming between neighboring cells (white arrowheads, Figure 2D). Furthermore, spontaneous contraction and calcium transients indicate functionality of iCMs (Figure 3A-C and Movie 1).
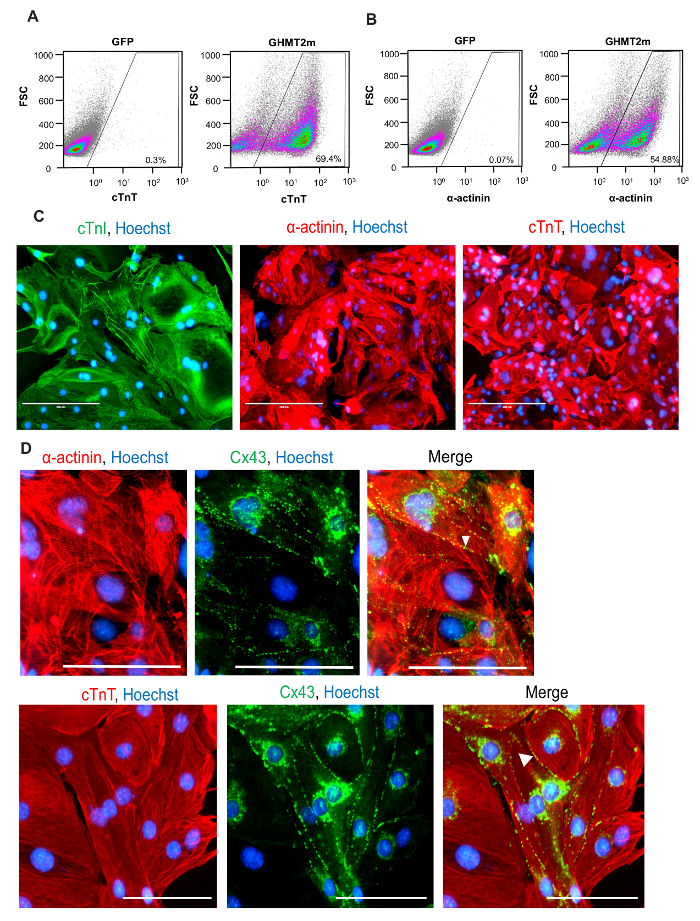
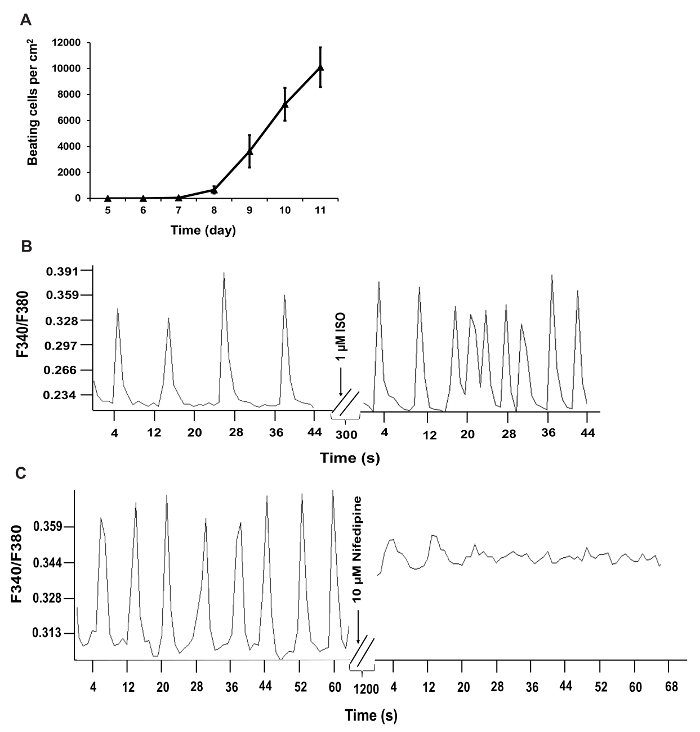
Figure 1: Timeline of Reprogramming and Optimal Plating of Cells. (A) Scatterplot of log-transformed RNA-seq data for MEFs generated in our lab versus ENCODE MEFs. The R2 value and linear regression line are shown. (B) Schematic of all critical steps in the GHMT2m-mediated reprogramming of MEFs. (C) PE cells at 70-80% confluence prior to transfection (top row, left panel) and GFP signal 48 hours post-transfection to indicate transfection efficiency (top row, middle and right panels). MEFs seeded sparsely prior to infection to prevent overcrowding over the time period of reprogramming (bottom row, left panel) and GFP signal 48 hours post-infection to indicate infection efficiency (bottom row, middle and right panels). Scale bar = 400 µM. Please click here to view a larger version of this figure.
Figure 2: Quantification and Characterization of Cardiac Protein Expression in iCMs.(A and B). Representative flow cytometry for cardiac Troponin T (A) and α-actinin (B) at Day 9 following GHMT2m transduction, n = 3. (C) ICC staining for cardiac markers at Day 14 following GHMT2m transduction. Green: cardiac Troponin I, Red: cardiac Troponin T (middle panel) and α-actinin (right panel), and Blue: Hoechst staining for nuclei. Representative images (n = 3). Scale bar = 200 µM (D) High magnification imagines of sarcomere structure (Red: cardiac α-actinin (top row) and cardiac Troponin T (bottom row)) and expression of gap junction protein Connexin 43 (green). White arrowheads indicate gap junctions formed between neighboring cells. Representative images (n = 3). Scale bar = 100 µM. Please click here to view a larger version of this figure.
Figure 3: Functional Quantification of iCMs.(A) Time course of beating cell counts following GHMT2m transduction. Beating cells were counted by eye and cell counts from 10 fields per dish over 3 dishes per experiment were averaged and included in panel A. (B and C) Recorded calcium transients of iCMs. Calcium transient frequency is altered in iCMs following treatment with 1 µM Isoproterenol or 10 µM nifedipine. F340/F380, the ratio of fluorescence intensity at 340 and 380 nm. Please click here to view a larger version of this figure.
 Movie 1: Beating iCMs. Movie showing beating iCMs at Day 12 in vitro. Please click here to view this video. (Right-click to download.)
Movie 1: Beating iCMs. Movie showing beating iCMs at Day 12 in vitro. Please click here to view this video. (Right-click to download.)
| iCM Media | ||
| Name of Reagent | Volume (mL) | Final Concentration |
| DMEM High Glucose | 320 | |
| Medium 199 | 80 | |
| Fetal Bovine Serum | 50 | 10% |
| Donor Horse Serum | 25 | 5% |
| MEM Essential Amino Acids, 50x | 10 | 1x |
| Sodium Pyruvate Solution, 100x | 5 | 1x |
| MEM Non-Essential Amino Acids, 100x | 5 | 1x |
| MEM Vitamin Solution, 100x | 5 | 1x |
| Insulin-Transferrin-Selenium, 100x | 5 | 1x |
| B27, 50x | 10 | 1x |
| Penicilin-Streptomycin | 5.5 | 1.1% |
| L-Glutamine supplement | 5.5 | 1.1% |
| PE Media | ||
| Name of Reagent | Volume (mL) | Final Concentration |
| DMEM High Glucose | 450 | |
| Fetal Bovine Serum | 50 | 10% |
| Penicilin-Streptomycin | 5.5 | 1.1% |
| L-Glutamine supplement | 5.5 | 1.1% |
| Blasticidin-HCl - 10mg/mL | 0.5 | 10 µg/mL |
| Puromycin dihydrochloride | 0.05 | 1 µg/mL |
| Growth Media | ||
| Name of Reagent/ Equipment | Volume (mL) | Final Concentration |
| DMEM High Glucose | 450 | |
| Fetal Bovine Serum | 50 | 10% |
| Penicilin-Streptomycin | 5.5 | 1.1% |
| L-Glutamine supplement | 5.5 | 1.1% |
Table 1: Culture Media. Recipes for culture medium used in this protocol.
| PE Cells | ||||||
| Cell culture dish | Surface Area (cm2) | Seeding Density (cells) | Media Volume (mL) | Total DNA per transfection (µg) | Transfection reagent (µL) | Opti-MEM (µL) |
| 15 cm | 176.7 | 11.25 x 106 | 20 | 36 | 108 | 1080 |
| 10 cm | 78.5 | 5 x 106 | 10 | 12 | 36 | 360 |
| 60 mm | 28.2 | 1.7 x 106 | 4 | 4 | 12 | 120 |
| 6 well plate | 9 | 0.54 x 106 | 2 | 1.3 | 3.9 | 39 |
| 12 well plate | 4 | 0.24 x 106 | 1 | 0.6 | 1.8 | 18 |
| 24 well plate | 2 | 0.12 x 106 | 0.5 | 0.3 | 0.9 | 9 |
| MEFs | ||||||
| Cell culture dish | Surface Area (cm2) | Seeding Density (cells) | Media Volume (mL) | Hexadimethrine bromide (µL) | ||
| 15 cm | 176.7 | 1.35 x 106 | 20 | 12 | ||
| 10 cm | 78.5 | 0.6 x 106 | 10 | 6 | ||
| 60 mm | 28.2 | 0.2 x 106 | 4 | 2.4 | ||
| 6 well plate | 9 | 0.1 x 106 | 2 | 1.2 | ||
| 12 well plate | 4 | 0.4 x 105 | 1 | 0.6 | ||
| 24 well plate | 2 | 0.2 x 105 | 0.5 | 0.3 |
Table 2: Seeding Densities for Reprogramming. Table of seeding densities for PE cells and MEFs for common cell culture dish formats.
Discussion
The present study outlines a high-efficiency strategy to directly reprogram fibroblasts into functional iCMs via delivery of GHMT2m reprogramming factors combined with suppression of pro-fibrotic signaling pathways. Using flow cytometry, immunofluorescent imaging, calcium imaging, and beating cell counts, we show the majority of cells in this protocol undergo successful reprogramming and adopt CM lineage fate. We have previously shown that the addition of anti-fibrotic compounds including the TGF-β type I receptor inhibitor A-83-01 yields an approximately 6-fold increase in the number of beating iCMs compared to transduction with GHMT2m alone, indicating that pro-fibrotic signaling is a strong endogenous barrier to cardiac reprogramming. This system can, therefore, be harnessed for drug screening to investigate mechanisms governing cardiomyogenesis; the addition of a small molecule alongside A-83-01 could uncover negative regulators of cardiomyogenesis. Conversely, the addition of a small molecule to GHMT2m alone could elucidate positive regulators of cardiomyogenesis. Unraveling the mechanisms through which these positive and negative regulators govern CM differentiation will result in a better understanding of cardiomyogenesis and lead to even more efficient reprogramming and maturation protocols.
Many variables affect reprogramming efficiency. We find that cell plating density greatly impacts both the production of retroviral particles in the case of PE cells, and the efficient uptake of all reprogramming factors in the case of MEFs. Furthermore, the passage number can affect these parameters as well. To ensure optimal production of viral particles, transfect PE cells before passage 20. To ensure efficient uptake of retroviral particles, do not passage MEFs after freezing.
Several recent studies have also demonstrated high-efficiency reprogramming of MEFs by knocking down PRC1 member Bmi along with expression of GMT30 or by overexpressing Akt in addition to GHMT37. Addition of ZFN281 to GHMT + Akt led to a 4-fold increase in troponin T positive cells in adult tail tip fibroblasts, attributed in part to suppression of inflammatory gene signatures38. Furthermore, the combination of a TGF-β signaling inhibitor and WNT signaling inhibitor increased GMT-mediated cardiac reprogramming39. Importantly, the signaling pathways involved in these studies do not appear to overlap. Therefore, it is likely that cross-talk between multiple signaling pathways could further enhance cardiac reprogramming.
A number of studies demonstrated reprogramming in vivo through the delivery of reprogramming factors in mice following left anterior descending (LAD) artery ligation25,32,39,40,41. These studies did show modest improvement in ventricular function following MI, underscoring the therapeutic potential of cardiac reprogramming on heart regeneration following injury. Our protocol reprograms MEFs with the highest efficiency in vitro to date. Therefore, it will be interesting to see if high-efficiency reprogramming is capable of fully restoring heart function post-injury in vivo. These studies could serve as a basis for translating this method into novel cell therapies for patients' post-infarction.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This research was supported by funds from the Boettcher Foundation's Webb-Waring Biomedical Research Program, American Heart Association Scientist Development Grant (13SDG17400031), University of Colorado Department of Medicine Outstanding Early Career Scholar Program, University of Colorado Division of Cardiology Barlow Nyle endowment, and NIH R01HL133230 (to K.S). A.S.R was supported by NIH/NCATS Colorado CTSA Grant Number TL1TR001081 and a pre-doctoral fellowship from the University of Colorado Consortium for Fibrosis Research & Translation (CFReT). This research was also supported by the Cancer Center Support Grant (P30CA046934), the Skin Diseases Research Cores Grant (P30AR057212), and the Flow Cytometry Core at the University of Colorado Anschutz Medical Campus.
References
- Benjamin EJ, et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation. 2017;136(20) doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE. Regenerating the heart. Nat. Biotechnol. 2005;23(7):845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: lessons from development. Genes & Development. 2011;25(4):299–309. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel EG, Braunwald E. A tale of coronary artery disease and myocardial infarction. N Engl J Med. 2012;366(1):54–63. doi: 10.1056/NEJMra1112570. [DOI] [PubMed] [Google Scholar]
- Packer M, et al. Effect of carvedilol on the morbidity of patients with severe chronic heart failure: results of the carvedilol prospective randomized cumulative survival (COPERNICUS) study. Circulation. 2002;106(17):2194–2199. doi: 10.1161/01.cir.0000035653.72855.bf. [DOI] [PubMed] [Google Scholar]
- The CAPRICORN Investigators. Effect of carvedilol on outcome after myocardial infarction in patients with left-ventricular dysfunction: the CAPRICORN randomized trial. The Lancet. 2001;357(9266):1385–1390. doi: 10.1016/s0140-6736(00)04560-8. [DOI] [PubMed] [Google Scholar]
- Marangou J, Paul V. Current attitudes on cardiac devices in heart failure: a review. Clin Ther. 2015;37(10):2206–2214. doi: 10.1016/j.clinthera.2015.08.012. [DOI] [PubMed] [Google Scholar]
- Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Bio. 2013;14(8):529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert KC, et al. Intracoronary autologous bone-marrow cell transfer after myocardial infarction: the BOOST randomized controlled clinical trial. Lancet. 2004;364(9429):141–148. doi: 10.1016/S0140-6736(04)16626-9. [DOI] [PubMed] [Google Scholar]
- Schachinger V, et al. Intracoronary bone marrow-derived progenitor cells in acute myocardial infarction. N. Engl. J. Med. 2006;355(12):1210–1221. doi: 10.1056/NEJMoa060186. [DOI] [PubMed] [Google Scholar]
- Huikuri HV, et al. Effects of intracoronary injection of mononuclear bone marrow cells on left ventricular function, arrhythmia risk profile, and restenosis after thrombolytic therapy of acute myocardial infarction. Eur. Heart J. 2008;29(22):2723–2732. doi: 10.1093/eurheartj/ehn436. [DOI] [PubMed] [Google Scholar]
- Janssens S, et al. Autologous bone marrow-derived stem-cell transfer in patients with ST-segment elevation myocardial infarction: double-blind, randomised controlled trial. Lancet. 2006;367(9505):113–121. doi: 10.1016/S0140-6736(05)67861-0. [DOI] [PubMed] [Google Scholar]
- Lunde K, et al. Intracoronary injection of mononuclear bone marrow cells in acute myocardial infarction. N. Engl. J. Med. 2006;355(12):1199–1209. doi: 10.1056/NEJMoa055706. [DOI] [PubMed] [Google Scholar]
- Hirsch A, et al. Intracoronary infusion of autologous mononuclear bone marrow cells or peripheral mononuclear blood cells after primary percutaneous coronary intervention: rationale and design of the HEBE trial - a prospective, multicenter, randomized trial. Am. Heart J. 2006;152(3):434–441. doi: 10.1016/j.ahj.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Hirsch A, et al. Intracoronary infusion of mononuclear cells from bone marrow or peripheral blood compared with standard therapy in patients after acute myocardial infarction treated by primary percutaneous coronary intervention: results of the randomized controlled HEBE trial. Eur. Heart J. 2011;32(14):1736–1747. doi: 10.1093/eurheartj/ehq449. [DOI] [PubMed] [Google Scholar]
- Karantalis V, et al. Autologous mesenchymal stem cells produce concordant improvements in regional function, tissue perfusion, and fibrotic burden when administered to patients undergoing coronary artery bypass grafting: The Prospective Randomized Study of Mesenchymal Stem Cell Therapy in Patients Undergoing Cardiac Surgery (PROMETHEUS) trial. Circ Res. 2014;114(8):1302–1310. doi: 10.1161/CIRCRESAHA.114.303180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Lian X, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci. 2012;109(27):E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, et al. Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell. 2016;166(2):451–467. doi: 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Yamanaka S. iPS cells: A source of cardiac regeneration. J. Mol. Cell. Cardiol. 2011;50(2):327–332. doi: 10.1016/j.yjmcc.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Shiba Y, et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature. 2016;538:388–391. doi: 10.1038/nature19815. [DOI] [PubMed] [Google Scholar]
- Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):357–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Katoku-Kikyo N, Keirstead SA, Kikyo N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc Res. 2013;100(1):105–113. doi: 10.1093/cvr/cvt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Berry EC, Fu J, Ieda M, Srivastava D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc. 2013;8(6):1204–1215. doi: 10.1038/nprot.2013.067. [DOI] [PubMed] [Google Scholar]
- Addis RC, et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifkovitz JL, Addis RC, Epstein JA, Gearhart JD. Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One. 2014;9(2):e89678. doi: 10.1371/journal.pone.0089678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell. 2016;18(3):382–395. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforou N, et al. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One. 2013;8(5):e63577. doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena TM, et al. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res. 2015;116(3):418–424. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena TM, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110(11):1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N, et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science. 2016;352(6290):1216–1220. doi: 10.1126/science.aaf1502. [DOI] [PubMed] [Google Scholar]
- Zhao Y, et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nature Communications. 2015;6:1–15. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Therapy. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Zhou H, Dickson ME, Kim MS, Bassel-Duby R, Olson EN. Akt1/protein kinase B enhances reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci. 2015;112(38):11864–11869. doi: 10.1073/pnas.1516237112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, et al. ZFN281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes & Dev. 2017;31:1770–1783. doi: 10.1101/gad.305482.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed TM, et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation. 2017;135:978–995. doi: 10.1161/CIRCULATIONAHA.116.024692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathison M, et al. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: Implications for clinical myocardial regeneration. J Thorac Cardiovasc Surg. 2017. pp. 329–339. [DOI] [PMC free article] [PubMed]


