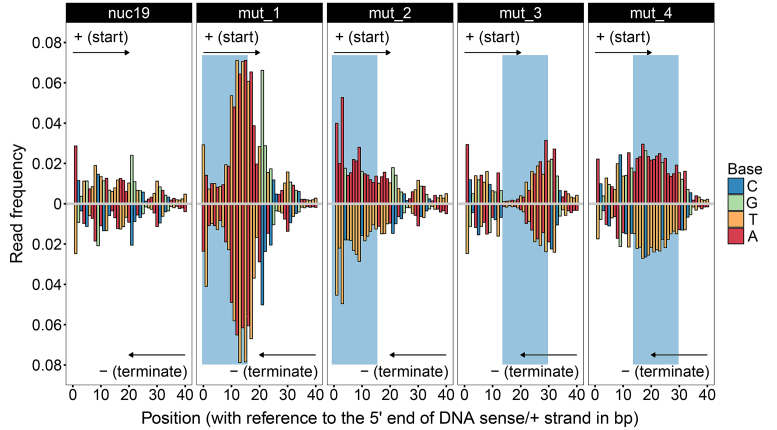
Figure 4.
MNase digestions on nuc19 and its four mutants. Read frequencies of nuc19 and its four mutants from 1-min digestion assays are shown. The digestion products were sequenced via paired-end sequencing approach. In ‘mut_1’ and ‘mut_2’, positions 0–15 on the ‘+’ strand of nuc19 were replaced with the TA-repeated and imperfect poly-A sequences, respectively. In ‘mut_3’ and ‘mut_4’, positions 14–29 on the ‘+’ strand of nuc19 were replaced with the TA-repeated and imperfect poly-A sequences, respectively. The mutated regions are shaded in blue. The frequencies of reads starting from each position in the range from 0 to 40 on the ‘+’ strand are shown in the upper panels. The frequencies of reads terminating at each position on the opposite strand (‘−’) of the mutated regions are shown in the lower panels. Note that frequencies at positions 0(+) and the paired positions on the opposite strand (‘−’) of nuc19, mut_2, mut_3 and mut_4 are above 0.08 and therefore not shown.