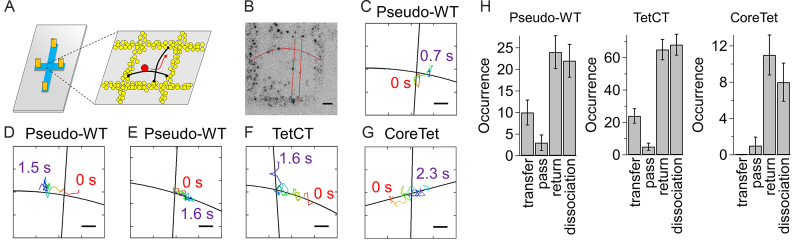
Figure 5.
Single-molecule observation of IST of p53. (A) Schematic drawing of the crisscrossed DNA garden. (Left) The crisscrossed flow cell. Orange cylinders, blue and gray denote connectors to the sample supply and disposal lines, the crisscrossed flow channel, and the coverslip, respectively. (Right) The enlarged view of the coverslip surface at the center of the flow channel. Yellow circles are neutravidin stamped on the coverslip, black curves are DNA, and black circles are biotin. The dynamics of ATTO532-p53 (red circle) near the intersection were observed by TIRF microscopy. (B) Representative fluorescence image of ATTO532-p53 observed on the crisscrossed DNA. Black dots represent single p53. Red curves are DNA strands obtained by fitting all the p53 coordinates sliding along DNA strands with a linear (vertical) and quadratic (horizontal) function. Scale bar in panel (B): 2 μm. (C)–(E) Representative trajectories of the pseudo-WT p53 near the intersection. Panels (C), (D) and (E) represent the images in which pseudo-WT p53 performed IST, passed over the intersection and dissociated from DNA at the intersection, respectively. Panels (F) and (G) represent the trajectories of the TetCT mutant performing IST and the CoreTet mutant which passed over the intersection, respectively. Black solid curves and rainbow curves in panels (C)–(G) denote DNA and the time course of trajectories in which red is the starting point, 0 s, respectively. Scale bars in panels (C)–(G): 0.5 μm. Panels (B)–(G) are the data obtained in the presence of 25 mM K+ and in the absence of Mg2+. (H) The observed numbers of events for p53 mutants entering the intersection. Errors were calculated by the bootstrap method with 1000 replicates (43).