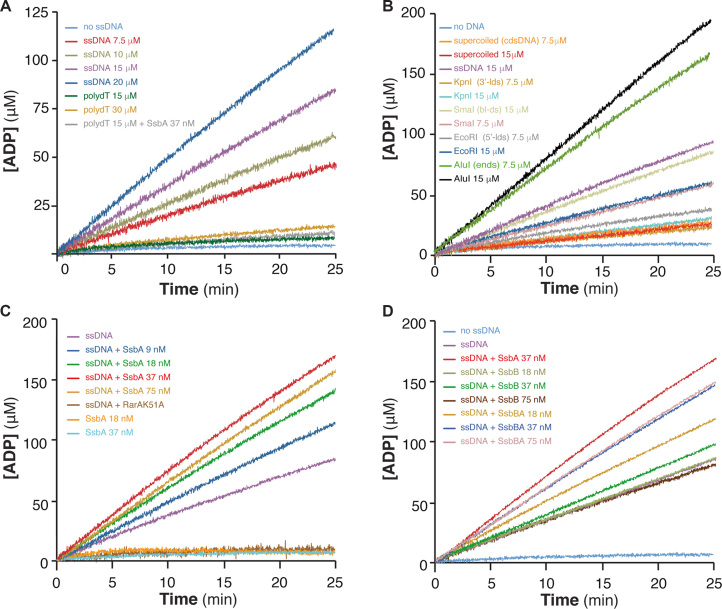
Figure 1.
RarA ATPase activity is stimulated by dsDNA-ssDNA junctions and dsDNA ends. (A) Stimulation by ssDNA containing secondary structures. RarA (25 nM) was incubated with increasing concentrations of circular 3199-nt ssDNA (7.5 to 20 μM) or linear 80-nt polydT ssDNA (15 and 30 μM) in buffer B containing 5 mM ATP, and the ATPase activity was measured (25 min, 37°C). The ATPase activity of RarA was also measured in the absence of ssDNA (no ssDNA, light blue) and in the presence of polydT and SsbA (gray). (B) Stimulation by dsDNA ends. RarA (25 nM) was incubated with two concentrations (7.5 and 15 μM) of various duplex DNA substrates: supercoiled 3199-bp dsDNA [cdsDNA], or dsDNA linearized with EcoRI [5′-ldsDNA], KpnI [3′-ldsDNA], SmaI [bl-dsDNA] or AluI [dsDNA-ends]. (C) Stimulation by SsbA bound to ssDNA. RarA (25 nM) was incubated with circular 3199-nt ssDNA (15 μM) and increasing concentrations of SsbA (9–75 nM). No ATPase activity is detected in the absence of ssDNA but presence of SsbA (18 nM and 37 nM, orange and blue) or when RarA was replaced by RarAK51A (ssDNA+K51A, dark brown). (D) ATPAse is stimulated by the C-terminal end of SsbA. RarA (25 nM) was incubated with circular 3199-nt ssDNA (15 μM) and increasing SsbB or SsbBA concentrations (18–75 nM). As a control, ATPase activity of RarA in the absence of ssDNA (no ssDNA, blue), in the presence of only ssDNA (magenta), or with ssDNA and SsbA (red) is shown. The amount of ATP hydrolysed was calculated as described (see Materials and Methods). Representative graphics are shown and quantification of the results are expressed as the mean ± SEM of >3 independent experiments (see Supplementary Table S2).