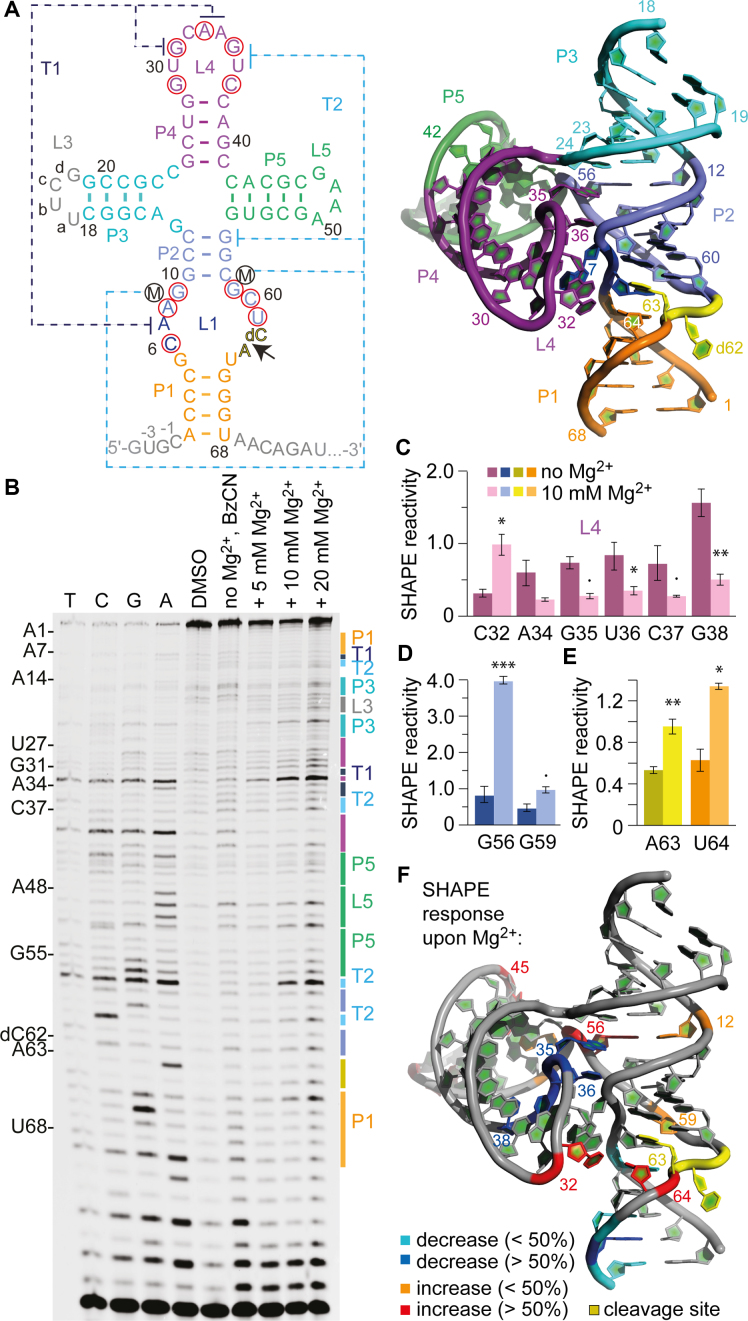
Figure 3.
SHAPE probing of the TS ribozyme. (A) Secondary structure representations of the TS RNA motif with 5′ and 3′ spacer sequences in gray. Red circles indicate highly conserved nucleosides. 3D structure (PDB accession number: 5Y87) using the same color code (right). (B) Typical gel for the probing of the pistol RNA structure with BzCN at 37°C. Lanes from left to right: T, C, G and A ladders, control in the absence of probing reagent, probing with BzCN, probing with BzCN and in the presence of 5, 10 and 20 mM MgCl2. Note that the fragment in the sequencing ladder that matches the size of the extension product defines the precise nucleotide that corresponds to +1 (therefore the color-coded stem-loop assignments at the right side of the gel is shifted +1 to nucleoside numbering on the left side). (C–E) Relative 2′-OH reactivity for selected bases of PK T1 obtained from quantification and normalization of the SHAPE probing results (mean of at least three independent experiments, error bars show standard deviation; two-sided paired t-test with *** P ≤ 0.001, ** P ≤ 0.01, * P ≤ 0.05, • P ≤ 0.1). For quantification of the SHAPE probing data for the complete TS RNA sequence see Supplementary Figure S7. (F) Projection of SHAPE reactivities of individual nucleotides on the 3D structure of the TS ribozyme (PDB accession number: 5Y87) following the color code as indicated in the legend.