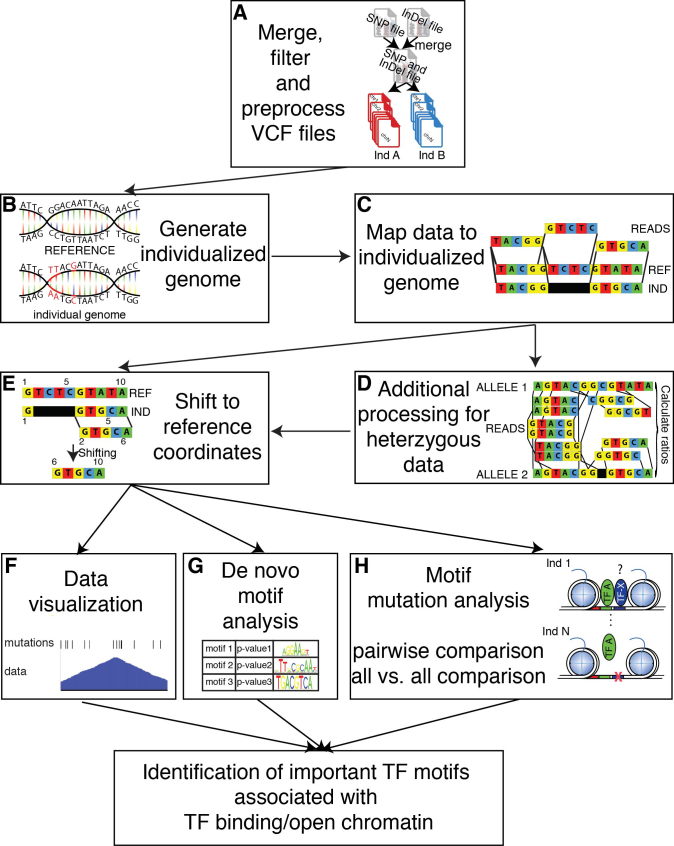
Figure 1.
Overview of the MMARGE pipeline. (A) MMARGE merges VCF files for SNPs and InDels, offers some basic filtering and split the merged VCF file into separate genotype-specific mutation files. (B) It then generates individual genomes by inserting the annotated mutations in the reference genome per genotype and (C) allows mapping of the experimental data sets to the individualized genomes. (D) The data mapped to the individualized genomes is then shifted back to the reference coordinates. (E) In case of heterozygous data additional processing is necessary. MMARGE offers (F) scripts for data visualization including BED files for genetic variation per genotype. It further offers (G) de-novo motif analysis for the individual genomes to make sure the enrichment analysis is performed on the correct sequence instead of the reference. MMARGE also offers a new algorithm (H) to associated TF binding motifs with genotype-specific binding for pairwise comparisons, as well as comparisons for many different individuals (all-versus-all comparison). Taken all of that together MMARGE is able to identify TF binding motifs that are functionally associated with TF binding.