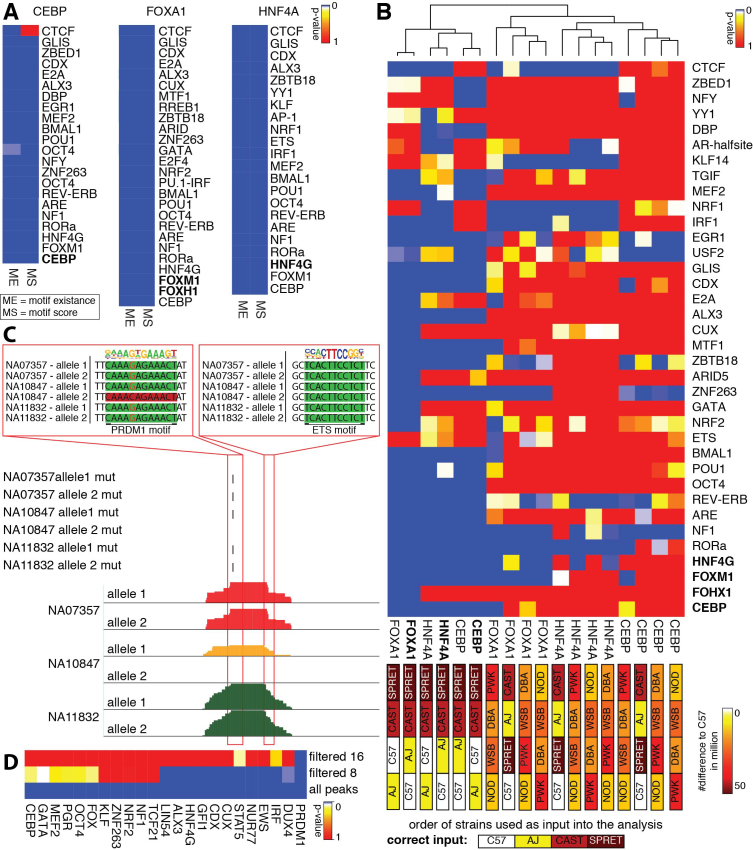
Figure 6.
Results of all-versus-all analysis. (A) Summary heat map of multiple testing corrected p-values of all-versus-all analysis of CEBP, FOXA1, and HNF4A ChIP-seq data sets from whole liver in AJ, C57, CAST, and SPRET. The analysis confirms the results from the pairwise analysis performed in Figure 5G. The same motifs are highly significance with slight variations independent of considering motif score (MS) or motif existence (ME). (B) Summary heat map of multiple testing corrected p-value of the all-versus-all analysis for CEBP, FOXA1, and HNF4A ChIP-seq data sets with the original order of the strains and shuffled order of the strains to assess sensitivity of MMARGE. The color of the boxes correlates to the number of mutations (from 0 – white to 50 million – brown). When very similar strains are switched (AJ and C57) the MMARGE results are clustered together. As soon as more diverse strains are switched or different strains are used, the results cluster as outliers to the original data and almost all motifs lose significance. (C) UCSC genome browser shot visualizing three human PU.1 datasets. The allele-specific loss of a PRDM1 motif close to an ETS motif causes allele-specific loss of PU.1 binding. (D) Summary heat map of multiple testing corrected p-values of transcription factor motifs significantly associated with PU.1 binding in human lymphoblastoid cell. Many TF motifs found to be significantly associated with PU.1 binding are either known to play important roles in B cell development and maintenance or cancer. By increasing the stringency of peaks included in the analysis (and decreasing the number of observations) the number of significant motifs decreases. Only PRDM1 is found as significant when using a filter of 16 reads.