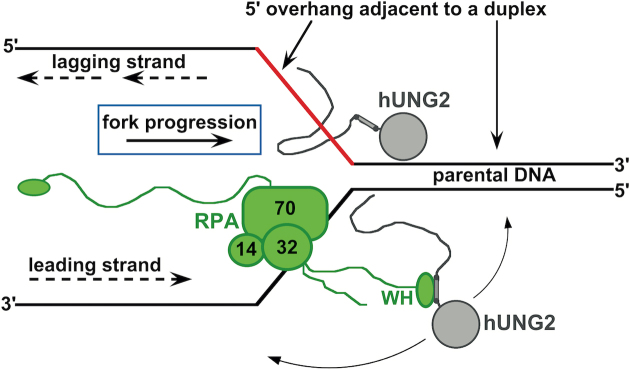
Figure 6.
Hypothetical targeting of hUNG2 at a simplified replication fork using its NTD. We propose that hUNG2 by itself could be targeted by transient protein-free ssDNA located on the 5′ end of a duplex region. This targeting could function to efficiently excise uracils prior to new strand synthesis. In the presence of RPA, which can bind to 5′ or 3′ ssDNA overhangs, we speculate that hUNG2 could be targeted to uracil bases before or after polymerase synthesis. When tethered to the winged-helix (WH) domain of RPA, hUNG2 can act on DNA strands in any direction. This arrangement could allow hUNG2 to preemptively remove uracils ahead of the fork, but could also allow hUNG2 to serve as a proofreading enzyme by acting on the newly synthesized strand. We note that other replication fork proteins (e.g. PCNA) were excluded for simplicity.