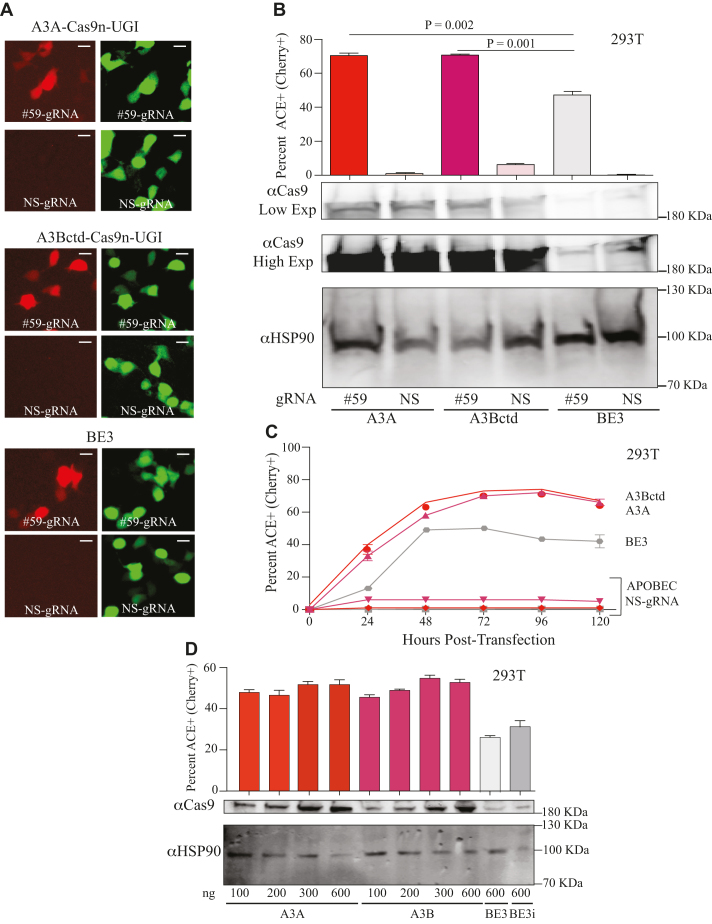
Figure 2.
High-efficiency editing by human A3A and A3Bctd editosomes. (A) Representative fluorescence microscopy images of ACE-activated, mCherry-positive 293T cells catalyzed by human A3A, human A3Bctd, or rat APOBEC1/BE3 editosomes (mCherry codon 59-directed gRNA versus NS-gRNA; inset white bar = 30 μm). (B) Quantification of the experiment in panel ‘A’ together with 2 independent parallel experiments (n = 3; average ± SD). The corresponding immunoblots of expressed APOBEC–Cas9n-UGI constructs are shown below (low and high exposures to help visualize BE3) with HSP90 as a loading control. (C) Time course of ACE activation in 293T cells catalyzed by human A3A, human A3Bctd, or rat APOBEC1/BE3 editosomes (mCherry codon 59-directed gRNA versus NS-gRNA; n = 3; mean ± SD; error bars smaller than symbols are not shown). (D) Titration data for 293T cells co-transfected with the ACE reporter, mCherry codon 59-directed gRNA, and different amounts of the indicated editosome constructs (100–600 ng; n = 3; mean ± SD). BE3i has an intron in the rat APOBEC1 portion of the construct, identical to the intron required for propagation of A3A and A3Bctd constructs in E. coli and for expression in mammalian cells.