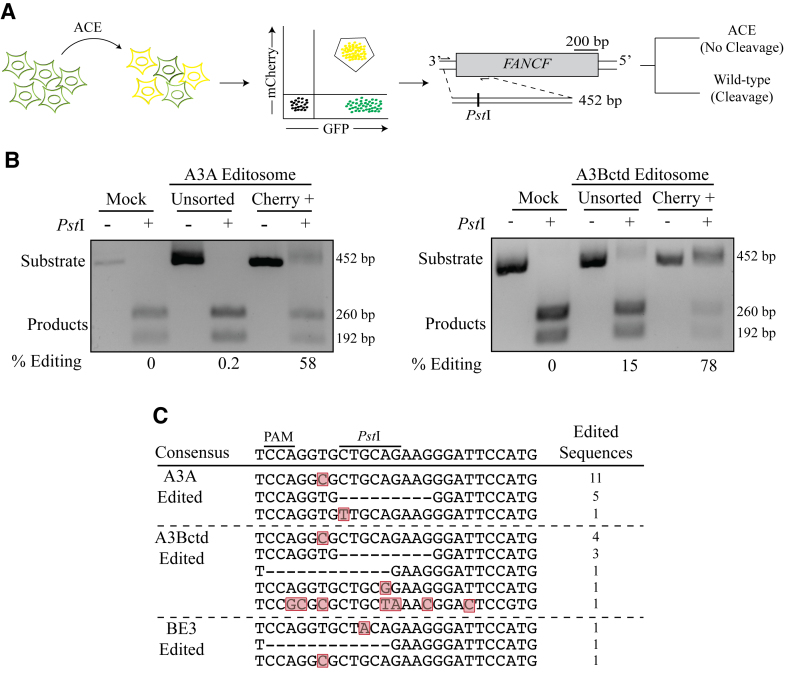
Figure 5.
ACE enriches for base-editing events at heterologous genomic loci. (A) Schematic of a co-transfection experiment resulting in ACE reporter activation (yellow shading represents mCherry and eGFP double-positive cells). FANCF and the PstI restriction assay used to quantify chromosomal base editing of this locus. Base editing events destroy the PstI cleavage site and block cleavage of the 452 bp amplicon into 260 and 192 bp products. (B) Representative agarose gels images showing the results of FANCF base editing by A3A and A3Bctd editsomes in 293T cells. The percentage of base editing was calculated by dividing the percentage of substrate band by the total of substrate and product bands following PstI cleavage for both unsorted and mCherry-positive cell populations. (C) Sanger sequencing results for the gRNA-binding region of the FANCF gene, which was recovered by high-fidelity PCR using genomic DNA from mCherry-positive 293T. Mutated nucleotides are highlighted in red and deleted nucleotides are indicated by hyphens. The number of times each sequence was recovered is indicated to the right.