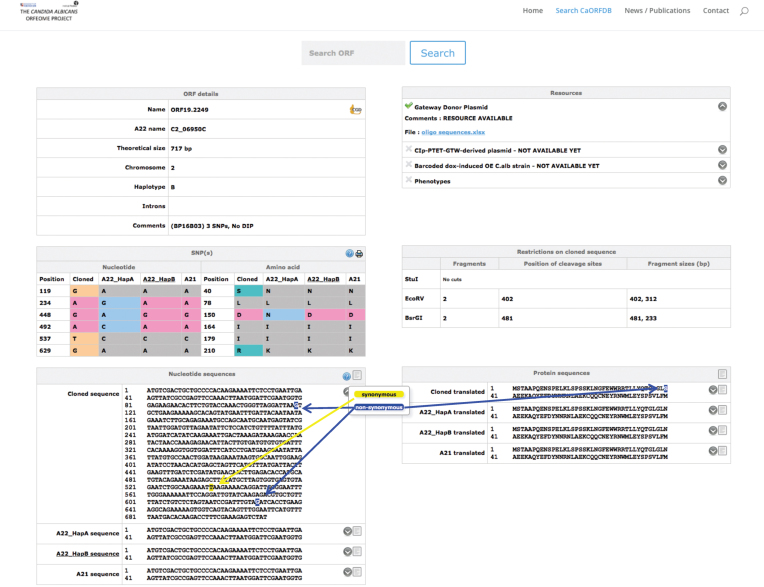
Figure 2.
Snapshot of the CandidaOrfDB interface. An example of an ORF page is shown. In the ‘ORF details’ box, the different ID names, the length and the chromosome location of the ORF of interest are displayed. The haplotype assigned to the cloned ORF is noted (A, B or A/B if there is no allelic differences or equal number of differences against both haplotypes). The coordinates of introns are indicated when present. The summary results of the sequence analysis against the reference sequence are presented. The ‘SNP(s)’ box shows a table that lists the sequence differences between the cloned ORF and the reference sequences (Haplotypes A and B from Assembly22, and Assembly21 sequences). The ‘Nucleotide and Protein sequences’ boxes display the sequences with a color code for synonymous and non-synonymous SNPs. All sequences can be downloaded. The ‘Resources’ box displays links towards information that is relevant to each resource, i.e. oligonucleotide sequences for the BP clones, barcode sequence for the overexpression plasmids and the C. albicans overexpression strains, position of the clone in the different plates of the collection, plasmid sequences. The ‘Restrictions on the cloned sequence’ box indicates the existence of restriction sites, as well as the size of the expected fragments for enzymes that are used in regards to subsequent applications of these donor plasmids.