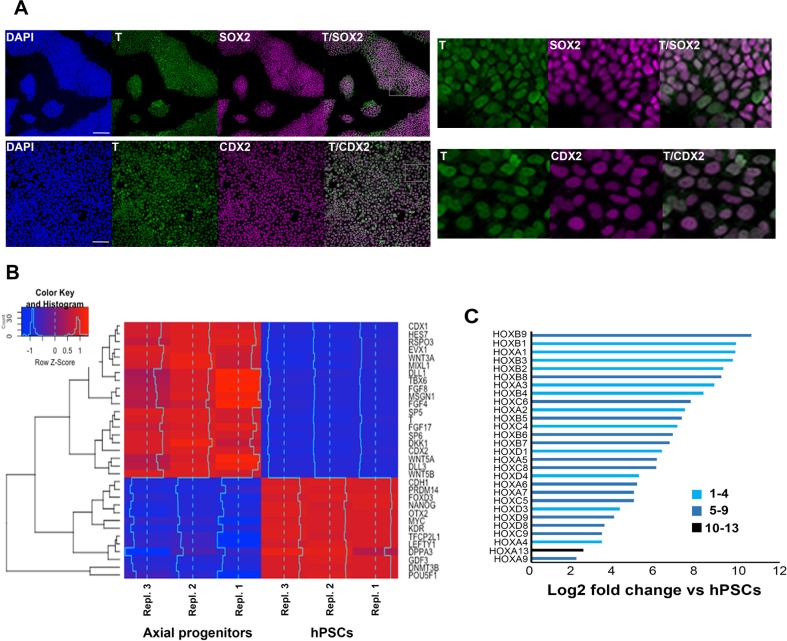
Figure 1. Transcriptome analysis of in vitro-derived human axial progenitors.
(A) Immunofluorescence analysis of expression of indicated markers in day 3 hPSC-derived axial progenitors. Magnified regions corresponding to the insets are also shown. Scale bar = 100 µm. (B) Heatmap showing the expression values of selected markers in three independent axial progenitor and hPSC sample replicates. The expression values (FPKM) were scaled to the row mean. The color key relates the heat map colors to the standard score (z-score). (C) Induction of all significantly upregulated HOX transcripts in axial progenitors relative to hPSCs. Paralogous HOX groups corresponding to different axial levels such as cervical (groups 1–4), brachial/thoracic (groups 5-9)and lumbosacral (groups 10-13) are indicated.