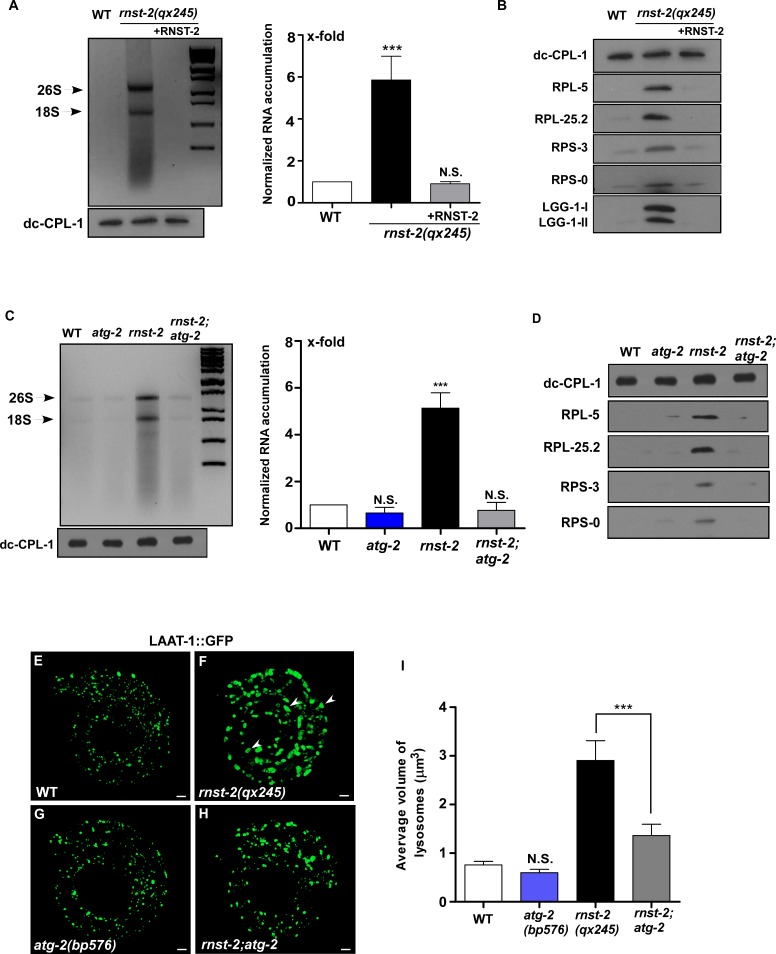
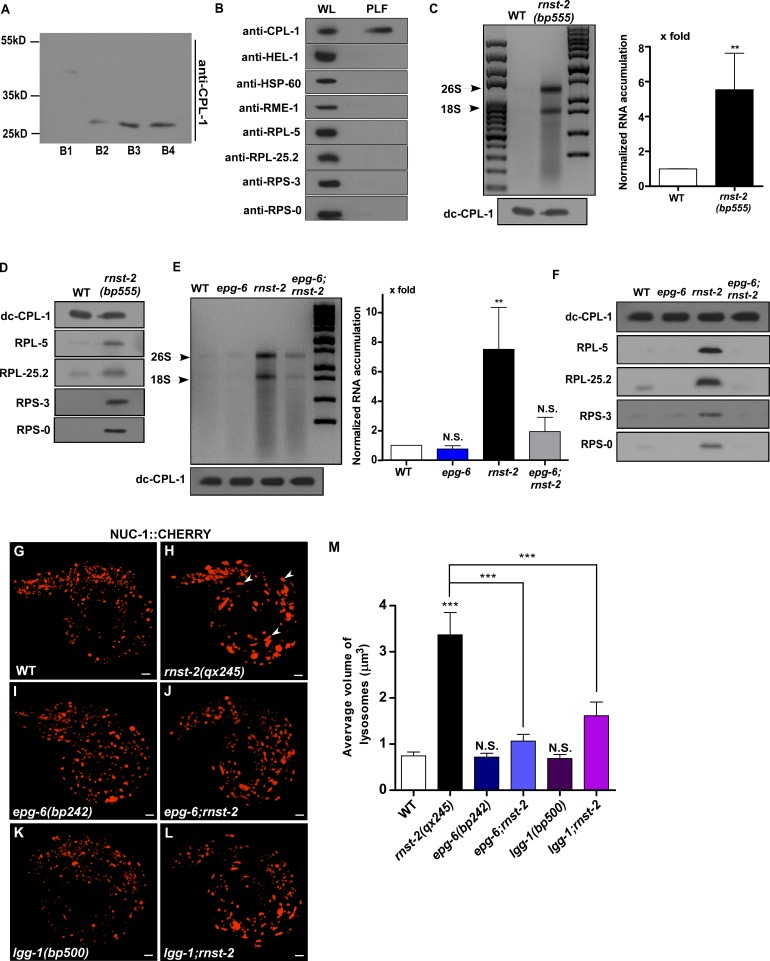
Figure 3. Loss of RNST-2 causes accumulation of rRNA and ribosomal proteins in lysosomes in an autophagy-dependent manner.
(A, C) RNA purified from lysosomes in the indicated strains was examined by agarose gel electrophoresis. Full processing of the lysosomal cathepsin CPL-1 (dc-CPL-1) was used to normalize the amount of lysosomes in each strain. The total extracted RNA was quantified and normalized to 1-fold in wild type (A, C, right panel). Abundant 26S and 18S rRNA was observed in lysosomes of rnst-2(qx245) mutants. At least three independent experiments were performed and data are shown as mean ± SD. (B, D) Accumulation of ribosomal proteins (large subunit: RPL-5, RPL-25.2; small subunit: RPS-3 and RPS-0) and LGG-1 in lysosomes was examined by western blot analysis in the indicated strains. Full processing of the lysosomal cathepsin CPL-1 (dc-CPL-1) was used to normalize the amount of lysosomes in each strain. (E–H) 3D reconstitution of the fluorescence images in 10–15 z-series (0.5 µm/section) in L1 larvae of the indicated strains expressing LAAT-1::GFP. Enlarged lysosomes (arrowheads) were observed in rnst-2(qx245). (I) Quantification of the average volume of lysosomes in the strains shown in (E–H). At least 10 worms were scored in each strain and data are shown as mean ± SD. In (A, C, I), one-way ANOVA with Tukey’s post hoc test was performed to compare all other datasets with wild type or datasets that are linked by lines (I). ***p<0.0001, N.S.: no significance.