Abstract
A dose responsive quantitative high throughput screen (qHTS) of >350,000 compounds against a human relaxin/insulin-like family peptide receptor (RXFP1) transfected HEK293 cell line identified 2-acetamido-N-phenylbenzamides 1 and 3 with modest agonist activity. An extensive structure-activity study has been undertaken to optimize the potency, efficacy, and physical properties of the series, resulting in the identification of compound 65 (ML-290), which has excellent in vivo PK properties with high levels of systemic exposure. This series, exemplified by 65, has produced first-in-class small-molecule agonists of RXFP1 and is a potent activator of antifibrotic genes.
Keywords: Relaxin, G-protein coupled receptor, antifibrotic, 2-acetamido-N-phenylbenzamide
Graphical Abstract
INTRODUCTION
The small-peptide hormone relaxin was discovered in 1926 and is primarily associated with pregnancy, due to its effects to relax pubic ligaments and soften the cervix to facilitate parturition.1,2 Since then it has been shown that blood concentrations of relaxin rise during the first trimester of pregnancy, stimulating cardiovascular and renal adjustments to accommodate the increased nutritional demands of the growing fetus and the elevated requirements for renal clearance of metabolic waste.3 Relaxin production increases cardiac output, arterial compliance, renal blood flow and a decrease in systemic vascular resistance during pregnancy.4–6 Both clinical and non-clinical studies using this hormone reinforce these cardiovascular effects in both males and females, which suggest the pharmacological utility of relaxin as a modulator of cardiovascular and renal functions in humans.7,8
The target of relaxin is a group of G-protein coupled receptors, relaxin/insulin-like family peptide receptor (RXFP).9,10 Some of the physiological effects of relaxin are mediated by its interaction with RXFP1, which modulates several signal transduction pathways.11,12 Relaxin activation of RXFP1 up-regulates the endothelin system which leads to vasodilation. Activation also promotes extracellular matrix remodeling through regulation of collagen deposition, proliferation, cell invasiveness, and perhaps most importantly, overall tissue homeostasis. It also moderates inflammation by reducing levels of inflammatory cytokines such as TNF-α and TGF-β, as well as the induction of angiogenesis via the activation of transcription of vascular endothelial growth factor (VEGF).13
Understanding the biological effects of RXFP1 activation by relaxin has led to the evaluation of relaxin as a pharmacologic agent for the treatment of patients with acute heart failure (AHF), preeclampsia, and hypertensive disease.6,14 A recent phase 3 clinical trial utilizing recombinant human relaxin-2 (serelaxin) met its primary endpoint of improving dysponea through the fifth day in patients admitted for acute heart failure.15,16 We have previously reported the identification and evaluation of a series of small-molecule human relaxin agonists.17,18 Optimized compounds from this series are potent and highly selective activators of RXFP1 with similar efficacy to the natural hormone in functional assays.19,20 Using quantitative high-throughput screening, over 350,000 compounds from our Molecular Libraries Small Molecule Repository (MLSMR) were screened in an effort to identify small-molecule agonists of RXFP1.21 This small molecule library was created by the NIH to combine chemical resources from both academic and private institutions to increase chemical space diversity of compounds for a large variety of screening campaigns. In this disclosure we present the syntheses, SAR studies, and compound optimization which led to the identification of preclinical candidate 65.
RESULTS AND DISCUSSION
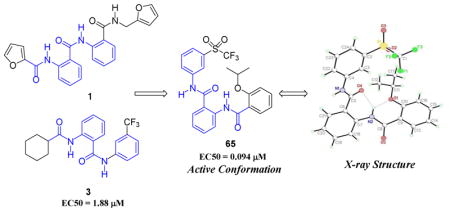
The ability of these compounds to increase cyclic adenosine monophosphate (cAMP) levels in a human RXFP1 transfected HEK293 cell line was measured, and two compounds (1 and 3), both having a unifying molecular motif of 2-acetamido-N-phenylbenzamide were discovered (Figure 1).17
Figure 1.
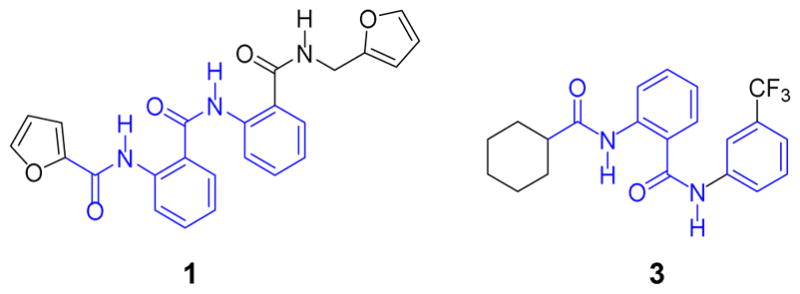
Initial RXFP1 hits from the Molecular Probe Center Network Library.
The activity is reported through two parameters: EC50 (concentration necessary to reach 50% of maximum cAMP signal) and maximum response (efficacy indicated as the maximum observed increase in cAMP response), which were used to perform SAR analysis. Efficacies were normalized to forskolin, which had an EC50 of 47 nM and a defined maximum response of 100%.22
Initial measurements of aqueous kinetic solubility of 1 and 3 (1.6 μg/mL, 3.7 μM and 0.9 μg/mL, 2.2 μM respectively) show poor solubility for these compounds. However, in assessing the stability of compounds 1 and 3 in mouse liver microsomes, compound 3 demonstrated very good stability after 60 minutes (70% parent remaining) versus 1 (2% parent remaining), so these data and the inherent problems associated with furans in mammalian metabolism led us to focus on the optimization of the cyclohexyl series based on compound 3.23 The general synthetic scheme to these is represented in Scheme 1.
Scheme 1.
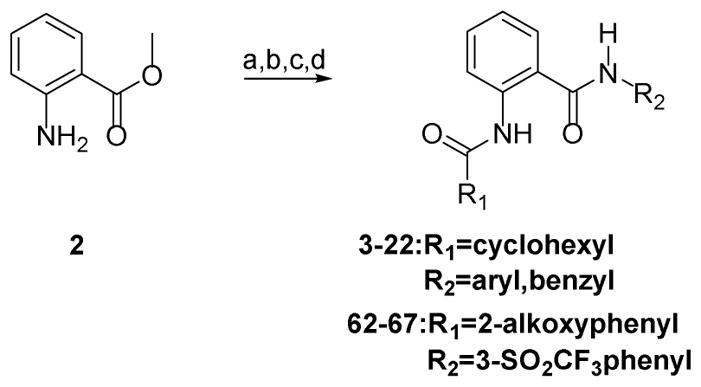
Synthesis of the Western Cyclohexyl and Optimized Analogues
Reagents and Conditions: For 3–22, 62–67: (a) acid chloride, DCM, TEA, 0 °C to RT, 1–3 h; (b) substituted aniline or benzylamine, 2M AlMe3 in toluene, 100 °C, 16 h; (c) For 17: 16, DCM, MCPBA, RT, 12h; (d) For 18: 13, phenylboronic acid, Pd(PPh3)4, 2M Na2CO3, DMF, microwave irrad., 100 °C, 1 h; For 19: 13, 2-(trifluoromethylphenyl)boronic acid, Pd(PPh3)4, 2M Na2CO3, DMF, microwave irrad., 100 °C, 1 h; For 20: 13, 3-(trifluoromethylphenyl)boronic acid, Pd(PPh3)4, 2M Na2CO3, DMF, μW., 100 °C, 1 h;
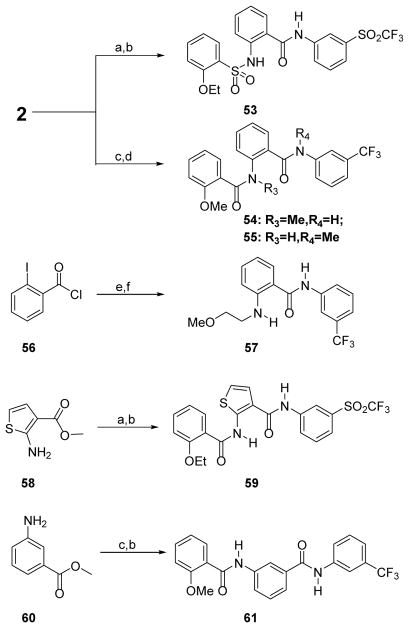
The first series of compounds 3–22 (Table 1) were prepared in a straightforward manner from 2 (see Experimental Standard Procedures), with the first amide formation performed by reaction with cyclohexylcarbonyl chloride followed by subsequent AlMe3 mediated second amide formation with the appropriate aniline or amine. This direct amidation using an aniline and an ester proved to be robust and broad in scope. Compounds 18–20 were prepared from bromo-compound 13 using standard Suzuki conditions.
Table 1.
SAR of the Cyclohexyl-2-Acetamido-N-Phenylbenzamide Series
| |||
|---|---|---|---|
| Cmpd | R | EC50(μM)a | Max. Responseb |
| 3 | 3-trifluoromethylphenyl | 1.88 | 92% |
| 4 | 4-trifluoromethylphenyl | 94.0 | 46% |
| 5 | phenyl | 94.0 | 57% |
| 6 | 2-methylphenyl | inactive | N/A |
| 7 | 3-methylphenyl | 37.4 | 65% |
| 8 | 4-methyphenyl | 187 | 32% |
| 9 | 3-tert-butlyphenyl | 2.65 | 70% |
| 10 | 3-nitrophenyl | 5.93 | 93% |
| 11 | 3-fluorophenyl | 13.3 | 81% |
| 12 | 3-chlorophenyl | 3.34 | 89% |
| 13 | 3-bromophenyl | 2.65 | 91% |
| 14 | 3-methoxyphenyl | 37.4 | 74% |
| 15 | 3-thiomethylphenyl | 5.29 | 84% |
| 16 | 3-trifluoromethythiophenyl | 1.88 | 90% |
| 17 | 3-trifluoromethylsulfonylphenyl | 1.06 | 87% |
| 18 | 3-biphenyl | inactive | N/A |
| 19 |
|
2.65 | 74% |
| 20 |
|
inactive | N/A |
| 21 |
|
inactive | N/A |
| 22 |
|
9.40 | 45% |
The EC50 is expressed in micromolar and is the concentration necessary to reach 50% of maximum cAMP signal.
The maximum response is the efficacy indicated as the maximum observed increase in cAMP response.
Table 1 discloses the initial results of our SAR studies. It is clear upon viewing the data that the 3-position of the pendant R-phenyl group provides the best agonism activity. Most of the compounds have single digit micromolar acivity, except for 7 (37.4 μM), 18 and 20 (both inactive). Interestingly, the 3-trifluoromethylbiphenyl derivative 19 is active (EC50 = 2.65 mM), leading us to believe that the trifluoromethyl moiety in this part of the molecule could be important by promoting the activation of RXFP1. This is further supported by compounds 16 and 17, the latter trifluoromethylsulfonyl compound being the most potent in this set, having an EC50 of 1.06 μM with an 87% of maximal activity. It is also worth noting that tert-butyl compound 9 maintains activity, and that less polar substituents may be preferred over more polar ones such as the 3-methoxy derivative 14.
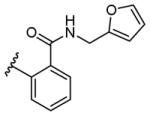
The unsubstituted phenyl compound 5 is minimally active as is 4-trifluoromethylsulfonyl phenyl compound 4. The inactivity of compound 6 might indicate that substituents at the ortho position impair agonism. Thus, either no substitution or 2- and 4-phenyl substitutions are deleterious to RXFP1 activation, with the 3-substituted phenyl compounds generally being more active. Drawing parallels to compounds 1 and 3, we combined the cyclohexyl and furanyl chemotypes, exemplified by compound 22, which retained some activity but did not encourage further investigation.
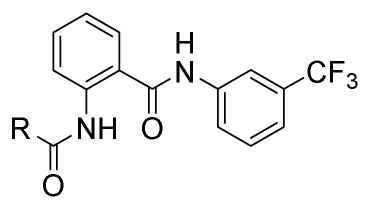
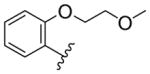
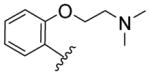
We then explored the replacement of the cyclohexyl group with various saturated alkyl rings, alkyl groups and differentially substituted aryl rings, while holding the 3-trifluoromethylphenyl eastern hemisphere region of the molecule constant (Scheme 2). Readily available BOC-protected anthranilic acid 23 was coupled to 3-trifluoromethylaniline and deprotected to afford 24a (which is also commercially available), and then converted to the amides 25–44 by reacting with the appropriate acid chloride, or coupled with the apprpriate carboxylic acid in the case of the cycloalkyl isosteres 27–30 (Table 2). The ethers 43–44 required an additional synthetic step using CuI catalyzed coupling of either 2-methoxyethan-1-ol or dimethylaminoethan-1-ol with iodo-compound 24b. It is evident from the data in Table 2 that the ring size of R is important. The cycloalkyl analogues 28–30 maintain good potency while the smaller cyclopropyl adduct 27 experiences a significant (> 4-fold) reduction in activity from the parent compound 3.
Scheme 2.
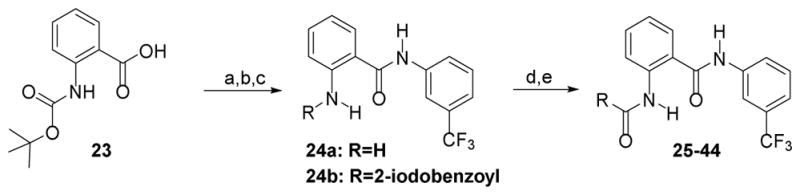
Synthesis of 3-Trifluoromethyl Analogues 25–44
Reagents and Conditions: (a) 3-trifluoromethylaniline, DCM, DMAP, EDC, 24 h; (b) TFA, DCM, 0 °C; (c) For 24b: 2-iodobenzoyl chloride, DCM, TEA, 0 °C, 1 h; (d) For 25–26, 31–42: aryl acid chloride, DCM, TEA, RT, 2 h; For 27–30: cycloalkyl carboxylic acid, DMF, EDC, DMAP, 12 h; (e) For 43: 24b, 2-methoxyethan-1-ol, Cs2CO3, CuI, 1,10-phenanthroline, toluene, sealed tube, 110 °C, 24 h; For 44: 24b, 2-dimethylaminoethan-1-ol, Cs2CO3, CuI, 1,10-phenanthroline, toluene, sealed tube, 110 °C, 24 h;
Table 2.
SAR of the Western Hemisphere N-3-Trifluorophenylbenzamide Series
| |||
|---|---|---|---|
| Cmpd | R | EC50 (μM)a | Max. Responseb |
| 25 | 3-pentyl | 5.29 | 59% |
| 26 | 1-butyl | 5.29 | 94% |
| 27 | cyclopropyl | 8.38 | 79% |
| 28 | cyclobutyl | 2.97 | 75% |
| 29 | cyclopentyl | 2.36 | 88% |
| 30 | cycloheptyl | 2.11 | 75% |
| 31 | benzyl | 8.38 | 86% |
| 32 | 3-pyridyl | 5.93 | 95% |
| 33 | 4-methoxyphenyl | 9.40 | 71% |
| 34 | phenyl | 1.32 | 64% |
| 35 | 3-methoxyphenyl | 1.88 | 94% |
| 36 | 2-methoxyphenyl | 0.334 | 99% |
| 37 | 2-methylphenyl | 4.20 | 90% |
| 38 | 2-chlorophenyl | 3.74 | 92% |
| 39 | 2-trifluoromethoxyphenyl | 1.18 | 96% |
| 40 | 2-ethoxyphenyl | 0.265 | 94% |
| 41 | 2-isopropoxyphenyl | 0.471 | 99% |
| 42 | 2,6-dimethoxyphenyl | 7.47 | 42% |
| 43 |
|
0.747 | 98% |
| 44 |
|
3.74 | 92% |
The EC50 is expressed in micromolar and is the concentration necessary to reach 50% of maximum cAMP signal.
The maximum response is the efficacy indicated as the maximum observed increase in cAMP response.
Compounds 27–30 clearly demonstrated a correlation between activity and ring size (3 < 4 < 5 < 6 > 7, EC50 = 8.38, 2.97, 2.36, 1.88, and 2.11 μM, respectively), with the 6-membered ring being the optimal size for activity. The branched and linear alkanes 25 and 26 are tolerated but not ideal for agonism activity. Again, as in compound 8, substitution at the 4-position substantially reduces the activity of compound 33 relative to 2-substitution (compound 36). The phenyl example 34 retains good activity while the 3-pyridyl 32 is an unfavorable substitution. A clear SAR profile begins to manifest as seen in the activity of micromolar to submicromolar 2-alkyl ethers 36, 39–41 and 43, with the 2-ethoxy compound 43 having the best activity at EC50 = 265 nM. These data were very encouraging as we demonstrated the additive effects of the 3-trifluorophenyl eastern hemisphere and the 2-alkoxyphenyl western hemisphere.
Imparting more solubility to this series of compounds would be desirable. Our next objective was to assess whether the central phenyl ring could tolerate derivatization in the form of pyridine analogues and retain, if not improve, potency and especially the solubility.
The four possible central core pyridine isomers were synthesized using readily available and inexpensive aminopyridines 45–48 (Scheme 3). From our SAR data the 3-sulfonylphenyl was chosen as a constant for the eastern region and 2-ethoxyphenyl was chosen for the western hemisphere. The resultant aminopyridines 49–52 were then assayed and the results are shown in Table 3. There is a substantial loss in activity of all but 3-pyridyl compound 51, which exhibits approximately a 3-fold reduction in activity from compound 40.
Scheme 3.
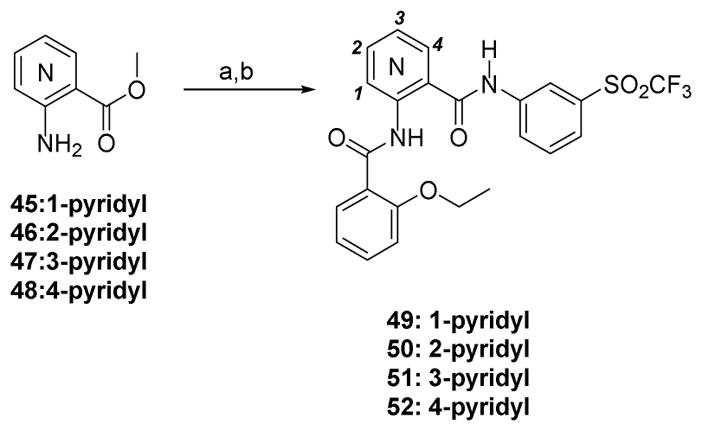
Synthesis of Pyridine Core Derivatives
Reagents and Conditions: (a) 2-ethoxybenzoylchloride, DCM, TEA, 0 °C-RT, 2 h, 11–94%; (b) 3-trifluoromethylsulfonylaniline, 2M Al-Me3 in toluene, 100 °C, 12 h, 56–75%;
Table 3.
SAR of Pyridine Central Core Derivatives
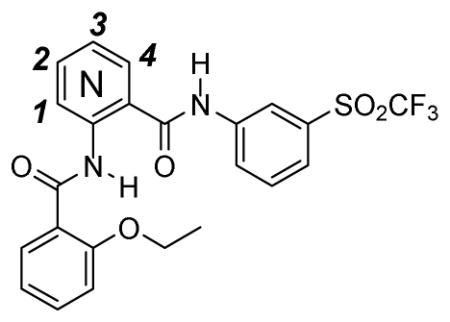
| ||||
|---|---|---|---|---|
| Cmpd | AC50 | Max. Response | PBS Solubilitya | RLM Stability (t1/2 min) |
| 49 (N1) | 1.67 | 98% | <1 mM | 19.85 |
| 50 (N2) | 2.97 | 99% | <1 mM | ND |
| 51 (N3) | 0.747 | 97% | ND | >30 |
| 52 (N4) | 4.71 | 73% | <1 mM | >30 |
Pion’s μSOL assay for kinetic solubility determination was used. Saturation shake-flask solubility method was adapted to a 96-well microtiter plate format and a co-solvent method with n-propanol as the reference compound was utilized.
The modification of the central benzamide linker was evaluated next, where the western region sulfonamide 53, N-methylated amides 54 and 55, amine 57, thiophene surragate 59, and 1,3-benzamide 61 were prepared using the standard chemistry detailed in this manuscript (Scheme 4), and then evaluated in the cAMP assay (Table 4). As our binding model predicts (vide infra), the 1,3 benzamide linked compound 61 was inactive, as were the N-methylated version 54 and sulfonamide 53. Methylated amide 55 retained weak activity, and surprisingly, so did the ethylmethoxyaniline 57. The thiophene isostere 59 had the best activity (EC50 = 2.35 μM).
Scheme 4.
Benzamide Linker Modifications: Sulfonamide, Amine, N-Methyl and 3-Acetamido-N-Phenylbenzamide Compounds
Reagents and Conditions:: (a) 2-ethoxyphenylsulfonylchloride, DCM, TEA, 0 °C-RT, 2 h; (b) 3-trifluoromethylsulfonylaniline, 2 M AlMe3 in toluene, 100 °C, 1–16 h, 10–82%; (c) 2-methoxybenzoylchloride, DCM, TEA, RT; For 54: (d) NaH, DMF, CH3I, 0 °C, 1.5 h, then (b); For 55: (b), then (d); (e) 3-trifluoromethylsulfonylaniline, DCM, TEA, 0 °C-RT; (f) 2-dimethylaminoethan-1-ol, Cs2CO3, CuI, 1,10-phenanthroline, toluene, sealed tube, 110 °C, 24 h;
Table 4.
SAR of Benzamide Linker Modifications
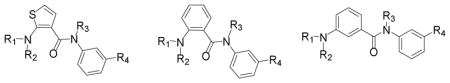
| |||||||
|---|---|---|---|---|---|---|---|
| Cmpd | R1 | R2 | R3 | R4 | Linker | EC50(μM)a | Max.b Response |
| 53 |
|
H | H | SO2CF3 |
|
inactive | N/A |
| 54 |
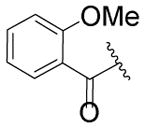
|
CH3 | H | CF3 |

|
inactive | N/A |
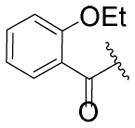
| 55 |

|
H | CH3 | CF3 |

|
29.7 | 77% |
| 57 |
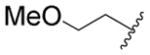
|
H | H | CF3 |

|
59.3 | 63% |
| 59 |
|
H | H | SO2CF3 |
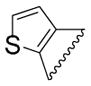
|
2.36 | 95% |
| 61 |

|
H | H | CF3 |
|
inactive | N/A |
The EC50 is expressed in micromolar and is the concentration necessary to reach 50% of maximum cAMP signal.
The maximum response is the efficacy indicated as the maximum observed increase in cAMP response.
A clear picture had emerged of the optimal SAR requirements for the 2-acetamido-N-phenylbenzamide series to effect RXFP1 activation. From the SAR data we determined that the 2-alkoxyphenyl western hemisphere and 3-trifluoromethylsulfonyl eastern hemisphere groups offered the best opportunity for discovering the most potent compounds, and gratifyingly, they did. Compounds 62–67 were synthesized according to the procedure in Scheme 1. The assay results of these compounds are shown in Table 5. The larger (65–67) or more hydrophobic alkyl groups (such as 64) confer the best activity, e.g., 62 or 63 (EC50 = 0.188 μM) as compared to compound 67 (EC50 = 0.047 μM). That the 3-trifluoromethylsulfonyl is a superior pharmacophore for activity to 3-trifluoromethyl on the eastern region phenyl is confirmed comparing 36 to 62 (EC50 = 0.334 μM versus EC50 = 0.188 μM), 40 to 63 (EC50 = 0.265 μM versus EC50 = 0.188 μM), and especially the obvious contrast between 41 and 65 (EC50 = 0.471 μM versus EC50 = 0.094 μM).
Table 5.
SAR of Optimized 2-Acetamido-N-Phenylbenzamide Compounds Including ML-290
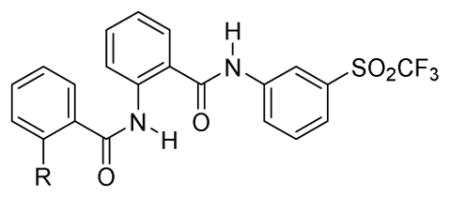
| |||||||
|---|---|---|---|---|---|---|---|
| Cmpd | R | EC50 (μM)a | Max. Responseb | ATP toxicity EC50 (μM)c | PBS Solubility (μM)d | Stability (t1/2 min) | |
| RLM | MLMe | ||||||
| 62 | OMe | 0.188 | 99% | 29.7 | 6.3 | >30 | ND |
| 63 | OEt | 0.188 | 99% | 18.8 | <1.1 | >30 | 1732 |
| 64 | OCH2CF3 | 0.067 | 97% | 29.7 | 3.3 | >30 | 100 |
| 65 | OCH(CH3)2 | 0.094 | 98% | 18.8 | 7.0 | 28.2 | 122 |
| 66 | OCH2CH2CH3 | 0.052 | 98% | 9.4 | 17.0 | 26.8 | 133 |
| 67 | OBu | 0.047 | 98% | 59.3 | 5.3 | 19.5 | 178 |
The EC50 is expressed in micromolar and is the concentration necessary to reach 50% of maximum cAMP signal.
The maximum response is the efficacy indicated as the maximum observed increase in cAMP response.
Cytotoxicity assay to measure the effect of compounds on cell viability by measuring ATP levels (ATPLite; Promega, Madison, WI).
Kinetic solubility analysis in PBS solution performed by Analiza (www.analiza.com).
Rodent microsomal stability analysis performed by Pharmaron (www.pharmaron.com).
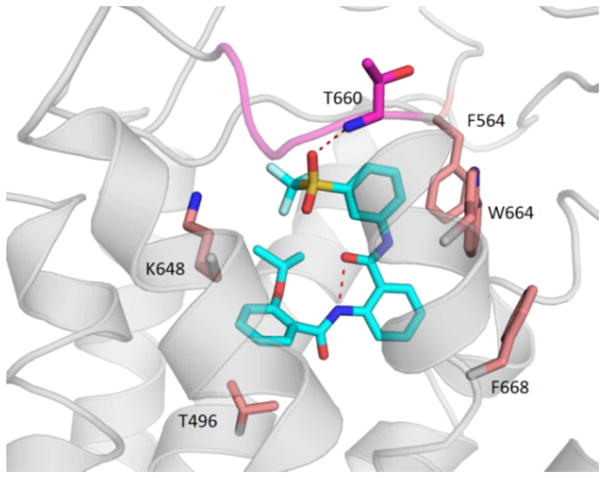
We have previously reported the structural elucidation and insight into the activation of the activation of human relaxin family peptide receptor by 65.18 Exhaustive site-directed mutagenesis in conjunction with computational modeling analysis allowed us to develop a predictive model of our small molecule agonists’ mode of binding. (Figure 2). Figure 2 shows the binding model of compound 65 (ML-290) to the RXFP1 receptor. The intramolecular hydrogen bonding interaction of the amide carbonyl to the threonine 660 is essential for favorable conformational bias within the active site. The inactivity of compound 54 and to a lesser extent the reduced activity of 55 (vide supra) clearly demonstrate that the replacement of the N-amido hydrogen with a methyl group results in the disruption of the required active site conformation for binding to RXFP1, as well as altering the critical T660 H-bonding interaction to one of the sulfonyl oxygens.
Figure 2.
Predicted model of ML290 bound in the TM binding site of hRXFP1. The receptor hRXFP1 is shown in ribbon (white) and key residues are shown in sticks (brown). In addition to an intramolecular H-bond, ML290 (sticks in cyan) forms a hydrogen-bond with the backbone N atom of residue T660 in ECL3 (magenta).18
The microsome stability, PBS solubility, and toxicity in both rat and mice were investigated for compounds 62–67. Rat and mouse microsome stability are excellent for these compounds (Table 5). An improvement in PBS solubility was also observed for compounds 62 and 64–67. It can be argued that the sulonyl group imparts greater solubility to these compounds as compared to the more lipophilic thrifluoromethyl moiety.
As we previously reported, 65 was chosen as our lead compound for further PK analysis because of favorable profiles which included: (1) selectivity against HEK293 cells transfected with the RXFP2 receptor (2) decreased adenosine triphosphate (ATP) cytotoxicity, and (3) especially its ability to activate the transcription of the known relaxin target gene VEGF in human monocyte leukemia cells (THP1). 9, 17
The pharmacokinetics of IV and PO delivery of 65 in mice was analyzed (Table 6). Compound 65 has an excellent half-life in plasma by both routes of administration and in heart by IV dosing, with the exposure in heart being about six times greater than plasma by IV administration. This is important as organ fibrosis (heart, liver, lung, etc) is an important indication of therapeutic intervention. While the plasma clearance is high, the high volume of distribution probably imparts the half-life observed. Importantly, no toxicity or abnormal behavior were detected. We have recently performed a maximum tolerated PO administration) dose study on C57/Bl6 mice (5 day with 5 day post-dosing observation), with no mortality observed at 90, 300, and 3000 mg/kg (see supplementary material). No obvious abnormalities were noted post-necropsy in any of the in vivo mammalian studies.
Table 6.
Pharmacokinetic Parameters for Compound 65 in Male C57/Bl6 Mice
| Route | Dosing Level (mg/kg) | CL obsa (mL/min/kg) | t1/2b(h) | C0c (μg/mL) | Cmaxd (μg/L) | AUClaste (μg-h/mL) | AUCinff (μg-h/mL) | MRTlastg (h) | Vss obsh (L/kg) | Fi (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| IV (plasma) | 3 | 67.2 | 6.6 | 0.49 | ND | 0.70 | 0.74 | 4.4 | 24.5 | ND |
| PO (plasma) | 30 | ND | 5.5 | ND | 0.30 | 0.98 | 1.00 | 3.1 | ND | 14 |
| IV (heart) | 3 | 8.78 | 6.3 | 2.93 | ND | 5.19 | 5.69 | 5.5 | 4.2 | ND |
| PO (heart) | 30 | ND | 1.0 | ND | 1.03 | 2.63 | 2.68 | 2.2 | ND | ND |
Total clearance after IV administration
Half-life
Concentration
Maximum serum concentration
Area under the curve
Area under the curve
Mean residense time
Volume of distribution in steady state
Bioavailability
Previously described antifibrotic effects of relaxin in various organs were associated with changes in transcriptional activity of key genes involved in extracellular matrix production and turnover.24 In fibrotic liver, hepatic stellate cells (HSCs) become activated and differentiate into profibrogenic myofibroblast-like cells. Importantly, RXFP1 gene expression is significantly upregulated in activated HSCs.25 It has been shown that relaxin treatment of activated HSCs leads to a significant decrease in profibrotic genes such as smooth muscle actin (ACTA2), transforming growth factor beta-1 (TGFB1), connective tissue growth factor (CTGF), and several collagens, but at the same time stimulates expression of matrix metalloproteinase 1 (MMP1) and peroxisome proliferator-activated receptor gamma (PPARG).26,27
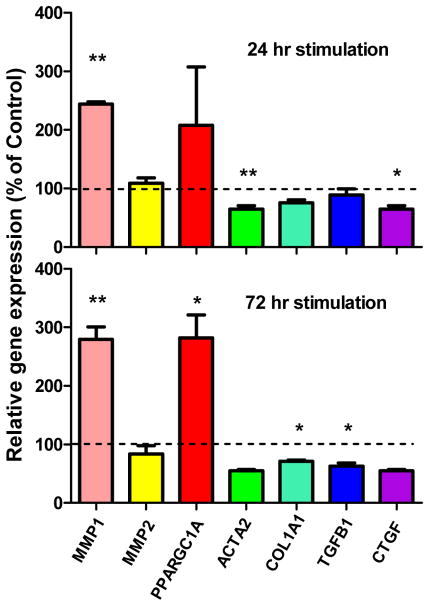
Therefore, we have tested the effect of ML290 on primary human HSC gene expression (Figure 3). Cells were cultured on plastic, which induces and maintains activation of HSCs.28 Upon treatment with 5μM of ML290, we observed a clear anti-fibrotic, relaxin-like gene expression pattern at 24 and 72 hr. Similar results have been recently reported by another group in the HSC LX-2 cell line using ML-290.29
Figure 3.
ML290 activates an antifibrotic gene expression profile in activated human hepatic stellate cells. HSCs were treated with 5μM of ML290 or DMSO for 24 and 72 hr. Gene expression was evaluated by quantitative RT-PCR, normalized to GAPDH gene expression and presented as the fold change vs DMSO-treated control. N=3, *p < 0.05, **p < 0.01.
CONCLUSION
Optimization of two lead compounds discovered by an extensive screening campaign of over 350,000 compounds at the National Center for the Advancement of Translational Science resulted in a series of potent, stable, selective and well-tolerated compounds which exhibit excellent systemic exposure in plasma and especially heart.
To summarize the SAR of the compounds presented here, the core aromatic ring must be ortho substituted to maintain activity. Replacing the central phenyl ring with either a thiophene or pyridine is tolerated although a decrease in activity is observed. The western hemisphere amide is critical for the activity.
For the western region, phenyl, pyridine and cycloalkanes of varying ring size are acceptable, but a six membered ring is optimal. An electron-donating substituent on the aromatic ring is tolerated at both the ortho- and meta-positions, with ortho-alkoxy having the best activity. At the eastern phenyl position, a meta-substitution is optimal and the trifluoromethylsulfonyl is best as a critical hydrogen bond interaction is formed between the sulfonyl oxygens and threonine 660.
Thus, the first small-molecule preclinical candidate agonist for the relaxin hormone receptor has been optimized through a medicinal chemistry approach and the resulting SAR is supported by a later-stage binding model obtained by iterative site-directed mutagenesis.
EXPERIMENTAL SECTION
Biology
Materials and Methods
Primary human hepatic stellate cells (HSCs) were grown on plastic in hepatic stellate growth media provided by the manufacturer (Zen-Bio Inc., Research Triangle Park, NC). Cells were seeded overnight in 35 mm poly-L-lysine-coated dishes (Corning, Durham, NC) at a density of 5×104 cells per dish and treated in triplicates with either 5μM of ML290 in DMSO (ML290 treatment) or DMSO (control) for 24 or 72 hr. Total RNA was extracted from cells samples with RNeasy kit (Qiagen, Germantown, MD). Quality and concentration of RNA was analyzed with a Nanovue (GE Healthcare Biosciences, Pittsburgh, PA). cDNA was synthesized using a Verso cDNA kit (Thermo Scientific, Fair Lawn, NJ). Quantitative PCR (qPCR) was performed using GoTaq qPCR Master Mix (Promega, Madison, WI), and gene specific primers in a RealPlex2 Mastercycler (Eppendorf, Westbury, NY) according to the manufacturer’s instructions. Values were normalized to GAPDH gene expression. The relative fold change in mRNA level was calculated by the comparative Ct (2−ΔΔC t) method. Results were analyzed using GraphPad Prism software (GraphPad Software, La Jolla, CA) for statistical significance using Student’s t test.
Measurements of cAMP concentration
cAMP assay was performed using HTRF cAMP HiRange kit (CisBio, Bedford, MA, USA). The THP1 and HEK293 cells (ATCC, Manassas, VA, USA) stably or transiently transfected with human RXFP1, RXFP2 or AVPR1B receptor were stimulated with relaxin, the compounds or forskolin for 30 min at 37 °C, 5% CO2, after which, 8 ml per well of each HTRF detection reagent (diluted according to assay kit directions in HTRF lysis buffer) was added.30,31 The plates were incubated for 30 min at room temperature, and the signal was read on a ViewLux (Perki-nElmer, Waltham, MA, USA) or a FLUOstar Omega (BMG Labtech, Cary, NC, USA) plate readers. Nonlinear regressions to the Hill equation were performed using Prism software (GraphPad Software, San Diego, CA, USA).
Modulation of HSC gene expression
Primary human hepatic stellate cells (HSCs) were grown on plastic in hepatic stellate growth media provided by the manufacturer (Zen-Bio Inc., Research Triangle Park, NC). Cells were seeded overnight in 35 mm poly-L-lysine-coated dishes (Corning, Durham, NC) at a density of 5×104 cells per dish and treated in triplicates with either 5μM of ML290 in DMSO (ML290 treatment) or DMSO (control) for 24 or 72 hr. Total RNA was extracted from cells samples with RNeasy kit (Qiagen, Germantown, MD). Quality and concentration of RNA was analyzed with a Nanovue (GE Healthcare Biosciences, Pittsburgh, PA). cDNA was synthesized using a Verso cDNA kit (Thermo Scientific, Fair Lawn, NJ). Quantitative PCR (qPCR) was performed using GoTaq qPCR Master Mix (Promega, Madison, WI), and gene specific primers in a RealPlex2 Mastercycler (Eppendorf, Westbury, NY) according to the manufacturer’s instructions. Values were normalized to GAPDH gene expression. The relative fold change in mRNA level was calculated by the comparative Ct (2–ΔΔCt) method. Results were analyzed using GraphPad Prism software (GraphPad Software, La Jolla, CA) for statistical significance using Student’s t test. The sequences of the primers are shown in Table 7.
Table 7.
Primer sequences used in qPCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GAPDHa | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCCc |
| ACTA2b | AAGCACAGAGCAAAAGAGGAAT | ATGTCGTCCCAGTTGGTGAT |
| COL1A1c | AGACAGTGATTGAATACAAAACCA | GGAGTTTACAGGAAGCAGACA |
| CTGFd | CCCAGACCCAACTATGATTAGAG | AGGCGTTGTCATTGGTAACC |
| MMP1e | GCACTGAGAAAGAAGACAAAGG | CTAAGTCCACATCTTGCTCTTG |
| MMP2f | CATACAGGATCATTGGCTACAC | TCACATCGCTCCAGACTTG |
| PPARGC1Ag | ATAAATCACACGGCGCTCTT | TGAGAGGGCCAAGCAAAG |
| TGFB1h | CACTCCCACTCCCTCTCTC | GTCCCCTGTGCCTTGATG |
Glyceraldehyde-3-phosphate dehydrogenase
Alpha-acti-2
Collagen
Connective tissue growth factor
Matrix metallopeptidase 1
Matrix metallopeptidase2
Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
Volume of distribution in steady state
Transforming growth factor beta-1
Chemistry
General Methods
All air or moisture sensitive reactions were performed under positive pressure of nitrogen with oven-dried glassware. Anhydrous solvents such as dichloromethane, N, N-dimethylformamide (DMF), acetonitrile, methanol and tetrahydrofuran were purchased from Sigma-Aldrich (St. Louis, Mo.). Preparative purification was performed on a Waters semi-preparative HPLC system (Waters Corp., Milford, Mass.). The column used was a Phenomenex Luna C18 (5 micron, 30 × 75 mm; Phenomenex, Inc., Torrance, Calif.) at a flow rate of 45.0 mL/min. The mobile phase consisted of acetonitrile and water (each containing 0.1% trifluoroacetic acid). A gradient of 10% to 50% acetonitrile over 8 minutes was used during the purification. Fraction collection was triggered by UV detection at 220 nM. Analytical analysis was performed on an Agilent LC/MS (Agilent Technologies, Santa Clara, Calif.). Method 1 (t1): A 7-minute gradient of 4% to 100% acetonitrile (containing 0.025% trifluoroacetic acid) in water (containing 0.05% trifluoroacetic acid) was used with an 8-minute run time at a flow rate of 1.0 mL/min. Method 2 (t2): A 3-minute gradient of 4% to 100% acetonitrile (containing 0.025% trifluoroacetic acid) in water (containing 0.05% trifluoroacetic acid) was used with a 4.5-minute run time at a flow rate of 1.0 mL/min. A Phenomenex Luna C18 column (3 micron, 3 × 75 mm) was used at a temperature of 50 °C. Purity determination was performed using an Agilent diode array detector for both Method 1 and Method 2. Mass determination was performed using an Agilent 6130 mass spectrometer with electrospray ionization in positive detection mode. 1H NMR spectra were recorded on Varian 400 MHz spectrometers (Agilent Technologies, Santa Clara, Calif.). Chemical shifts are reported in ppm with undeuterated solvent (DMSO-d6 at 2.49 ppm) as internal standard for DMSO-d6 solutions. All of the analogs tested in the biological assays have a purity of greater than 95% based on both analytical methods. High resolution mass spectrometry was recorded on Agilent 6210 Time-of-Flight (TOP) LC/MS system. Confirmation of molecular formula was accomplished using electrospray ionization in the positive mode with Agilent Masshunter software (Version B.02).
General Protocol A
A solution of methyl or ethyl benzoate (0.191 mmol) and amine (0.383 mmol) in toluene (2.00 mL) was treated at room temperature with AlMe3 (0.192 mL, 2.0 M in toluene, 0.384 mmol). The reaction mixture was stirred overnight at 100° C and then quenched with 100 μL of water. The mixture was concentrated, redissolved in 2.00 mL of DMSO, filtered and purified via C18 reverse phase HPLC to give the final product.
General Protocol B
A solution of carboxylic acid (0.178 mmol) in DMF (2.00 mL) was treated at room temperature with 2-amino-N-(3-(trifluoromethyl)pheny1)benzamide (25.0 mg, 0.089 mmol) followed by EDC (17.l mg 0.089 mmol) and DMAP (10.9 mg, 0.089 mnol). The reaction mixture was stirred overnight at room temperature. The mixture was purified via C18 reverse phase HPLC to give the final product.
General Protocol C
A solution of 2-amino-N-(3- (trifluoromethyl)phenyl)benzamide (50.0 mg, 0.178 mmol) in dichloromethane (2.00 mL) and TEA (0.075 mL, 0.535 mmol) was treated at room temperature with carbonyl chloride (0.357 mmol). The reaction mixture was stirred at room temperature for 2 h. The mixture was concentrated, re-dissolved in 2.00 mL of DMSO, filtered and purified via C18 reverse phase HPLC to give the final product.
General Protocol D
A solution of carboxylic acid (0.357 mmol) in DMF (2.00 mL) was added DIPEA (0.093 mL, 0.535 mmol) and HATU (136 mg, 0.357 mmol). The reaction mixture was stirred at room temperature for 5 min, followed by 2-amino-N-(3-(trifluoromethyl)-phenyl)benzamide (50.0 mg, 0.178 mmol). The reaction mixture was stirred overnight at room temperature, filtered and purified via C18 reverse phase HPLC to give the final product.
General Protocol E
A mixture of 2-bromo-N-(3- (trifluoromethyl)phenyl)benzamide (50.0 mg, 0.145 mmol, 1.0 equiv.) or N-(3-bromophenyl)-2-(cyclohexanecarboxamido)benzamide (100 mg, 0.249 mmol, 1.0 equiv.), boronic acid or pinacol ester (2.0 equiv.) and Pd(PPh3)4 (0.05 eq) in DMF (1.50 mL) and 2.0 N Na2CO3 (0.50 mL) aqueous solution was heated in μW at 100° C for 30 min to 1 h. The reaction was cooled to room temperature, and a small portion of Si-THIOL was added to remove palladium. The reaction mixture was filtered and purified via C18 reverse phase HPLC to give the final product.
General Protocol F
2-Iodo-N-(3-(trifluoromethyl) phenyl)benzamide (100 mg, 0.256 mmol), amine (0.767 mmol), and CuCl (7.59 mg, 0.077 mmol) in DMF (1.00 mL) was stirred at room temperature for 15–30 min. The reaction was treated with a small portion of Si-THIOL to remove palladium, filtered and purified via C18 reverse phase HPLC to give the final product.
General Protocol G
A solution of thio-compound (0.255 mmol) in dichloromethane (3.00 mL) was treated at room temperature with MCPBA (220 mg, 1.27 mmol). The reaction mixture was stirred over-night at room temperature. 10% Aqueous NaHSO3 solution was added to quench excess MCPBA and the mixture was stirred at room temperature for 15 min. The reaction mixture was partitioned between dichloromethane and water. The organic layer was separated, dried (Na2SO4), filtered, concentrated, and purified via C18 reverse phase HPLC to give the final product.
General Protocol H
A tube was charged with CuI (0.1 equiv.), 1,10-phenanthroline (0.2 equiv.), Cs2CO3 (2.0 equiv.), iodo substrate (1.0 equiv.) and alcohol (2.0 equiv.) in toluene (2.00 mL) under N2. The tube was sealed and the reaction mixture was stirred at 110 ° C for 24 h. The resulting mixture was cooled to room temperature and treated with a small portion of Si-THIOL to remove copper. The mixture was concentrated, dissolved in 2.00 mL of DMSO, filtered and purified via C18 reverse phase HPLC to give the final product.
3-(Cyclohexanecarboxamido)-N-(3-(trifluoromethyl)phenyl)-benzamide (3)
The title compound was prepared according to general protocol A. Yield: 35%; LC-MS Retention Time: t1 (Method 1) = 6.516 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.53 (s, lH), 10.00 (s, lH), 8.21 (t, J = 2.l Hz, lH), 8.14 (t, J = 2.0 Hz, lH), 8.03 (ddd, J = 8.2, 1.2, 1.0 Hz, lH), 7.81 (ddd, J = 8.l, 2.2, 1.1 Hz, lH), 7.55–7.64 (m, 2H), 7.37–7.49 (m, 2H), 2.24–2.41 (m, lH), 1.69–1.92 (m, 4H), 1.57–1.69 (m, lH), 1.08–1.50 (m, 5H); 19F NMR (376 MHz, DMSO- d6) δ ppm −61.29 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 174.40, 167.65, 140.12, 138.19, 132.20, 130.29, 129.90, 129.58, 129.19, 125.92, 125.07, 124.52, 123.62, 122.48, 120.58, 120.54, 117.14, 117.10, 45.47, 29.45, 25.82, 25.50; HRMS (ESI) m/z (M+H) calcd. for C21H22F3N2O2, 391.1628; found 391.1637.
2-(Cyclohexanecarboxamido)-N-(4-(trifluoromethyl)phenylbenzamide (4)
The title compound was prepared according to general protocol A. Yield; 62%; LC-MS Retention Time: t1 (Method 1) = 6.943 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.65 (s, lH), 10.29 (s, lH), 8.09 (dd, J =. 8.4, 1.0 Hz, lH), 7.91 (d, J = 8.6 Hz, 2H), 7.74 (dd, J =7.8, 1.4 Hz, lH), 7.71 (d, J = 8.4 Hz, 2H), 7.5l (ddd, J = 8.4, 7.3, 1.6 Hz, lH), 7.21 (td, J =7.6, 1.2 Hz, lH), 2.18–2.35 (m, lH), 1.80 (dd, J =l2.7, 2.9 Hz, 2H), 1.68 (dt, J = l2.2, 3.4 Hz, 2H), 1.59 (d, J = 11.9 Hz, lH), 1.01–1.43 (m, 5H); 19 FNMR (376 MHz, DMSO-d6) δ ppm −60.26 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 173.94, 167.32, 142.64, 141.12, 137.79, 134.12, 131.86, 131.13, 128.90, 125.96, 123.64, 123.51, 122.03, 120.34, 45.10, 29.02, 25.41, 25.13. HRMS (ESI) m/z (M+H) calcd. For C21H22F3N20, 391.1628; found 391.1628.
2-(Cyclohexanecarboxamido)-N-phenylbenzamide (5)
The title compound was prepared according to general protocol A. Yield: 68% LC-MS Retention Time: t1 (Method 1) 6.326 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.54 (s, lH), 10.37 (s, lH), 8.23 (dd, J =8.2, l.0 Hz, lH), 7.77 (dd, J =7.9, l.5 Hz, lH), 7.63–7.71 (m, 2H), 7.49 (ddd, J = 8.4, 7.2, 1.6 Hz, lH), 7.28–7.41(m, 2H), 7.19 (td, J =7.6, 1.3 Hz, lH), 7.07–7.14 (m, lH), 2.17–2.34 (m, lH), 1.76–1.88 (m, 2H), 1.69 (ddd, J = l2.5, 3.4, 3.2 Hz, 2H), l.59 (d, J = 12.l Hz, lH), 1.03–1.43 (m, 5H); 13C NMR (151 MHz, DMSO-d6) δ 176.98, 170.13, 141.77, 141.35, 134.88, 131.88, 131.73, 127.14, 126.60, 125.99, 124.48, 123.91, 48.42, 43.00, 42.86, 42.72, 42.58, 42.44, 42.30, 42.16, 32.14, 28.48, 28.20; HRMS (ESI) m/z (M+H) calcd. For C20H23N2O2, 323.l754; found 323.l758.
2-(Cyclohexanecarboxamido)-N-o-tolylbenzamide (6)
The title compound was prepared according to general protocol A. Yield: 38%; LC-MS Retention Time: t1 (Method 1) = 6.401 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.00 (s, lH), 10.09 (s, lH), 8.38 (dd, J = 8.4, 1.2 Hz, lH), 7.91 (d, J = 7.8 Hz, lH), 7.51 (ddd, J =8.6, 7.2, 1.7 Hz, lH), 7.25–7.35 (m, 2H), 7.10–7.25 (m, 3H), 2.21 (s, 3H), 2.16–2.28 (m, lH), 1.77–1.91 (m, 2H), 1.69 (dt, J = l2.4, 3.5 Hz, 2H), l.53–l.65 (m, lH), 0.99–1.44 (m, 5H); 13C NMR (101 MHz, DMSO-d6) δ 173.86, 167.36, 139.15, 135.79, 134.00, 132.11, 130.36, 128.60, 126.87, 126.43, 126.11, 122.63, 121.31, 120.78, 45.61, 29.06, 25.36, 25.06, 17.81; HRMS (ESI) m/z (M+H) calcd. for C21H25N2O2, 337.1911; found 337.1912.
2-(Cyclohexanecarboxamido)-N-m-tolylbenzamide (7)
The title compound was prepared according to general protocol A. Yield: 55%; LC-MS Retention Time: t1 (Method 1) = 6.632 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.54 (s, lH), 10.30 (s, lH), 8.24 (dd, J =8.3, 1.1 Hz, lH), 7.76 (dd, J =7.8, 1.4 Hz, lH), 7.38–7.54 (m, 3H), 7.10–7.31 (m, 2H), 6.93 (dddd, J = 7.5, l.5, l.l, 0.8 Hz, lH), 2.29 (s, 3H), 2.19–2.3S (m, lH), 1.82 (dd, J = l2.8, 2.6 Hz, 2H), 1.69 (dt, J = l2.4, 3.4 Hz, 2H), l.S9 (d, J = l2.3 Hz, lH), 1.04–1.42 (m, 5H); 13C NMR (101 MHz, DMSO-d6) δ 173.88, 167.00, 138.53, 138.30, 137.75, 131.75, 128.74, 128.45, 124.76, 123.40, 122.83, 121.39, 121.32, 118.09, 45.33, 29.05, 25.38, 25.07, 21.19; HRMS (ESI) m/z (M+H) calcd. for C21H25N2O2, 337.1911; found 337.1908.
2-(Cyclohexanecarboxamido)-N-p-tolylbenzamide (8)
The title compound was prepared according to general protocol A. Yield: 49%; LC-MS Retention Time: t1 (Method 1) = 6.636 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.61 (s, 1H), 10.30 (s, 1H), 8.26 (dd, J =8.2, 0.8 Hz, 1H), 7.77 (dd, J = 7.8, 1.4 Hz, 1H), 7.52–7.59 (m, 2H), 7.48 (ddd, J =8.4, 7.2, 1.6 Hz, 1H), 7.18(dd, J = 7.8, 1.2 Hz, 1H), 7.11–7.17 (m, 2H), 2.26 (s, 3H), 2.16–2.36 (m, 1H), 1.82 (dd, J = 13.0, 2.1 Hz, 2H), 1.69 (ddd, J =12.2, 3.3, 3.1 Hz, 2H), l.51–1.64 (m, 1H), 1.02–1.47 (m, 5H); 13C NMR (101 MHz, DMSO-d6) δ 174.29, 167.36, 138.85, 136.50, 133.57, 132.20, 129.45, 129.14, 123.60, 123.23, 121.67, 121.37, 45.84, 29.50, 25.83, 25.54, 20.98; HRMS (ESI) m/z (M+H) calcd. for C21H25N2O2, 337.1911; found 337.1916.
N-(3-(tert-Butyl)phenyl)-2-(cyclohexanecarboxamido)benzamide (9)
The title compound was pre-pared according to general protocol A. Yield: 80%; LC-MS Retention Time: t1 (Method 1) = 7.324 min; 1H NMR (400 MHz, DMSO-d6) δ 10.51 (s, 1H), 10.30 (s, 1H), 8.20 (d, J = 8.3 Hz, 1H), 7.77 (d, J = 7.7 Hz, 1H), 7.61 (d, J = 8.4 Hz, 2H), 7.54 – 7.45 (m, 1H), 7.27 (dd, J = 8.3, 7.6 Hz, 1H), 7.24 – 7.11 (m, 2H), 2.30 – 2.20 (m, 1H), 1.91 – 1.79 (m, 2H), 1.69 (d, J = 12.7 Hz, 3H), 1.60 (d, J = 13.3 Hz, 1H), 1.43 – 1.32 (m, 4H), 1.27 (s, 9H); 13C NMR (101 MHz, DMSO-d6) δ 173.82, 167.22, 140.35, 137.77, 132.75, 131.81, 130.31, 128.73, 124.21, 123.55, 123.04, 121.80, 120.03, 118.95, 45.10, 29.00, 25.37, 25.05, 45.46, 33.89, 30.55, 29.44, 25.82, 25.51; HRMS (ESI) m/z (M+H) calcd. For C24H31N2O2, 379.2380; found 379.2387.
2-(Cyclohexanecarboxamido)-N-(3-nitrophenyl)benzamide (10)
The title compound was prepared according to general protocol A. Yield: 30%; LC-MS Retention Time: t1 (Method 1) = 6.392 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.76 (s, lH), 10.26 (s, lH), 8.69 (t, J = 2.2 Hz, lH), 8.01–8.l5 (m, 2H), 7.95 (ddd, J = 8.3, 2.3, 1.0 Hz, lH), 7.75 (dd, J = 7.8, 1.6 Hz, lH), 7.64 (t, J = 8.2 Hz, lH), 7.46–7.56 (m, lH), 7.22 (td, J = 7.6, 1.2 Hz, lH), 2.19–2.35 (m, lH), 1.79 (dd, J = 12.5, 2.5 Hz, 2H), 1.68 (dt, J = 3.1 Hz, 2H), 1.58 (d, J = 12.5 Hz, lH), 1.03–1.43 (m, 5H); 13C NMR (101 MHz, DMSO-δ6) δ 174.40, 167.74, 148.30, 140.58, 138.17, 132.30, 130.49, 129.23, 126.87, 125.04, 123.66, 122.55, 118.72, 115.03, 45.46, 29.44, 25.82, 25.51; HRMS (ESI) m/z (M+H) calcd. for C20H22N3O4, 368.1605; found 368.1616.
2-(Cyclohexanecarboxamido)-N-(3-fluorophenyl)benzamide (11)
The title compound was prepared according to general protocol A. Yield: 73%; LC-MS Retention Time: t1 (Method 1) = 6.529 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.51 (s, lH), 10.33 (s, lH), 8.12 (dd, J = 8.3, 1.1 Hz, lH), 7.73 (dd, J = 7.7, l.5 Hz, lH), 7.64 (dt, J = ll.7, 2.3 Hz, lH), 7.43–7.55 (m, 2H). 7.37 (td, J = 8.2. 6.7 Hz, lH), 7.20 (td, 7.5, 1.2 Hz, lH), 6.82–6.98 (m, lH), 2.21–2.35 (m, lH), 1.81 (d, J = l1.5 Hz, 2H), 1.69 (dt, J = 12.4, 3.3 Hz, 2H), 1.59 (d, J = 11.7 Hz, lH), 1.02–1.43 (m, 5H); 19F NMR (376 MHz, DMSO-d6) δ ppm −112.20–112.29 (m, 1F); HRMS (ESI) m/z (M+H) calcd. for C20H22FN2O2, 341.1660; found 341.1660.
N-(3-Chlorophenyl)-2-(cyclohexanecarboxamido)benzamide (12)
The title compound was prepared according to general protocol A. Yield: 40%; LC-Retention Time: t1 (Method 1) = 6.898 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.48 (s, lH), 10.31 (s, lH), 8.11 (d, J = 7.8 Hz, lH), 7.85 (t, J = 2.0 Hz, lH), 7.71(dd, J = 7.7, 1.3 Hz, lH), 7.59 (dd, J = 8.0, 1.8 Hz, lH), 7.43–7.54 (m, lH), 7.35 (t, J = 8.l Hz, lH), 7.06–7.24 (m, 2H), 2.26 (tt, J = 11.3, 3.6 Hz, lH), 1.79 (d, J = 14.l Hz, 2H), 1.68 (ddd, J = 12.7, 3.0, 2.7 Hz, 2H), 1.52–1.62 (m, lH), 1.03–1.41 (m, 5H); 13C NMR (101 MHz, DMSO-d6) δ 173.90, 167.10, 140.30, 137.91, 132.85, 131.77, 130.27, 128.73, 124.17, 123.55, 123.04, 121.80, 120.03, 118.95, 45.10, 29.00, 25.37, 25.05; HRMS (ESI) m/z (M+H) calcd. for C20H22ClN2O2, 357.1364; found 357.1366.
N-(3-Bromophenyl)-2-(cyclohexanecarboxamido)benzamide (13)
The title compound was prepared according to general protocol A. Yield: 90%; LC-MS Retention Time: t1 (Method 1) = 6.904 min; 1 H NMR (400 MHz, DMSO-d6) δ ppm 10.47 (s, lH), 10.31 (s, lH), 8.11 (dd, J = 8.4, 1.0 Hz, lH), 7.94–8.04 (m, lH), 7.72 (dd, J = 7.8, 1.6 Hz, lH), 7.64 (ddd, J = 6.7, 2.3, 2.2 Hz, lH), 7.49(ddd, J = 8.4, 7.2, 1.6 Hz, lH), 7.24–7.35 (m, 2H), 7.19 (td, J = 7.6, 1.2 Hz, lH), 2.21–2.37 (m, lH), 1.80 (d, J = 11.5 Hz, 2H), 1.69 (dt, J = l2.2, 3.2 Hz, 2H). l.59 (d, J = 12.5 Hz, lH), 1.07–1.42 (m, 5H); 13C NMR (151 MHz, DMSO-d6) δ 177.00, 170.14, 143.70, 141.05, 134.85, 133.69, 131.84, 129.67, 129.53, 127.55, 126.17, 125.07, 124.93, 122.38, 48.30, 32.24, 28.61, 28.29; HRMS (ESI) m/z (M+H) calcd. for C20H22BrN2O2, 401.0859; found 401.0859.
2-(Cyclohexanecarboxamido)-N-(3-methoxyphenyl)benzamide (14)
The title compound was pre-pared according to general protocol A. Yield: 47%; LC-MS Retention Time: t1 (Method 1) = 6.344 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.47 (s, lH), 10.33 (s, lH), 8.20 (dd, J = 8.4, 1.0 Hz, lH), 7.74 (dd, J = 7.8, 1.4 Hz, lH), 7.49 (ddd, J = 8.4, 7.2, 1.6 Hz, lH), 7.33 (t, J = 2.2 Hz, lH), 7.13–7.30 (m, 3H), 6.69 (ddd, J = 7.8, 2.5, 1.4 Hz, lH), 3.73 (s, 3H), 2.26 (tt, J =11.3, 3.6 Hz, lH), 1.82 (dd, J = 12.5, 2.7 Hz, 2H), 1.69 (ddd, J = 12.4, 3.1, 2.8 Hz, 2H), l.5l–1.64 (m, 1H), 1.02–1.41 (m, 5H); 13C NMR (101 MHz, DMSO-d6) δ 174.31, 167.47, 159.82, 140.28, 138.58, 132.17, 129.84, 129.16, 124.20, 123.34, 121.95, 113.48, 109.81, 107.12, 55.49, 45.72, 29.48, 25.83, 25.53; HRMS (ESI) m/z (M+H) calcd. for C21H25N2O3, 353.1860; found 353.1856.
2-(Cyclohexanecarboxamido)-N-(3-(methylthio)phenyl)benzamide (15)
The title compound was prepared according to general protocol A. Yield: 36%; LC-MS Retention Time: t11 (Method 1) = 6.687 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.44 (s, lH), 10.37 (s, lH), 8.18 (d, J = 8.2 Hz, lH), 7.75 (dd, J = 7.8, 1.6 Hz, lH), 7.63 (t, J = 2.l Hz, lH), 7.44–7.55 (m, 2H), 7.29 (t, J = 7.9 Hz, lH), 7.20 (td, J = 7.6, 1.3 Hz, lH), 7.01 (ddd, J = 7.8, 2.0, 1.0 Hz, lH), 2.46 (s, 3H), 2.19–2.35 (m, lH), 1.76–1.90 (m, 2H), 1.65–1.76 (m, 2H), 1.52–1.65 (m, lH), 1.03–1.47 (m, 5H); 13C NMR (101 MHz, DMSO-d6) δ 173.89, 167.05, 139.25, 138.41, 138.08, 131.74, 129.12, 128.70, 123.86, 122.95, 121.60, 121.35, 117.88, 117.23, 45.23, 29.04, 25.38, 25.08, 14.67; HRMS (ESI) m/z (M+H) calcd. for C21H25N2O2S, 369.1631; found 369.1625.
2-(Cyclohexanecarboxamido)-N-(3-(trifluoromethyl)phenyl)benzamide (16)
The title compound was prepared according to general protocol A. Yield: 50%; LC-MS Retention Time: t1 (Method 1) = 7.349 min; 1H NMR (600 MHz, DMSO-d6) δ 10.58 (s, 1H), 10.28 (s, 1H), 8.12 (d, J = 2.0 Hz, 1H), 8.04 (d, J = 8.2 Hz, 1H), 7.90 (ddd, J = 8.3, 2.2, 1.1 Hz, 1H), 7.72 (dd, J = 7.8, 1.6 Hz, 1H), 7.50 (q, J = 7.2, 6.5 Hz, 2H), 7.43 (dt, J = 7.9, 1.3 Hz, 1H), 7.21 (td, J = 7.6, 1.2 Hz, 1H), 2.26 (tt, J = 11.5, 3.6 Hz, 1H), 1.82 – 1.75 (m, 2H), 1.67 (dt, J = 12.9, 3.5 Hz, 2H), 1.58 (dt, J = 13.0, 3.4 Hz, 1H), 1.37 – 1.17 (m, 5H), 1.11 (dddd, J = 16.1, 12.4, 7.9, 3.5 Hz, 1H), 1.02 (s, 2H); 13C NMR (151 MHz, DMSO-d6) δ 177.05, 170.21, 143.28, 140.76, 134.83, 134.25, 133.79, 133.33, 131.88, 131.75, 130.35, 127.94, 126.33, 126.18, 125.19, 48.41, 32.09, 28.48, 28.18; HRMS (ESI) m/z (M+H) calcd. for C21H22F3N2O2S, 423.1349; found 423.1360.
2-(Cyclohexanecarboxamido)-N-(3((trifluoromethyl)sulfonyl)phenyl)benzamide (17)
The title compound was prepared according to general protocol G. Yield: 80%; LC-MS Retention Time: t1 (Method 1) = 6.964 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.83 (s, lH), 10.16 (s, lH), 8.57 (t, J =l.6 Hz, lH), 8.26 (ddd, J = 5.7, 3.7, 2.2 Hz, lH), 7.93 (dd, J = 8.3, 1.3 Hz, lH), 7.76–7.86 (m, 2H), 7.72 (dd, J = 7.8, 1.6 Hz, lH), 7.52 (ddd, J = 8.5, 7.2, 1.6 Hz, lH), 7.23 (td, J = 7.6, 1.2 Hz, lH), 2.16–2.36 (m, lH), 1.73–1.87 (m, 2H), 1.62–1.73 (m, 2H), 1.50–1.62 (m, lH), 0.82–1.46 (m, 5H); 19F NMR (376 MHz, DMSO-d6) δ ppm −78.42 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 174.04, 167.31, 141.09, 137.28, 131.77, 131.30, 129.82, 128.79, 128.47, 125.80, 125.24, 123.52, 122.71, 122.61, 120.99, 120.55, 118.38, 116.22, 44.85, 29.00, 25.42, 25.11; HRMS (ESI) m/z (M+H) calcd. for C21H22F3N2O4S, 455.1247; found 455.1253.
N-([1,1′-Biphenyl]-3-yl)-2-(cyclohexanecarboxamido) benzamide (18)
The title compound was pre-pared according to general protocol A. Yield: 39%; LC-MS Retention Time: t1 (Method 1) = 7.181 min; 1H NMR (600 MHz, DMSO-d6) δ 10.51 (d, J = 13.2 Hz, 2H), 8.22 (dd, J = 8.4, 1.1 Hz, 1H), 8.00 (t, J = 2.0 Hz, 1H), 7.82 (dd, J = 7.9, 1.6 Hz, 1H), 7.73 (dt, J = 8.0, 1.6 Hz, 1H), 7.67 – 7.61 (m, 2H), 7.56 – 7.36 (m, 6H), 7.23 (td, J = 7.6, 1.2 Hz, 1H), 2.29 (tt, J = 11.5, 3.6 Hz, 1H), 1.88 – 1.81 (m, 2H), 1.71 (dp, J = 10.7, 3.5 Hz, 2H), 1.64 – 1.57 (m, 1H), 1.37 (qd, J = 12.5, 3.3 Hz, 2H), 1.26 (qt, J = 12.6, 3.3 Hz, 2H), 1.15 (qt, J = 12.5, 3.5 Hz, 1H); 13C NMR (151 MHz, DMSO-d6) δ 177.02, 170.21, 143.70, 143.15, 142.36, 141.23, 134.87, 132.36, 132.11, 131.85, 130.73, 129.72, 126.97, 126.08, 125.49, 124.68, 122.92, 122.25, 48.36, 32.16, 28.48, 28.18; HRMS (ESI) m/z (M+H) calcd. for C26H27N2O2, 399.2067; found 399.2071.
2-(Cyclohexanecarboxamido)-N-(2′-(trifluoromethyl)-[1, l′-biphenyl]-3- yl)benzamide (19)
The title compound was prepared according to general protocol E. Yield: 25%; LC-MS Retention Time: t1 (Method 1) = 7.417 min; 1H NMR (600 MHz, DMSO-d6) δ 10.50 (s, 1H), 10.41 (s, 1H), 8.16 (dd, J = 8.2, 1.1 Hz, 1H), 7.85 (dd, J = 7.9, 1.2 Hz, 1H), 7.82 – 7.71 (m, 4H), 7.63 (td, J = 7.6, 1.1 Hz, 1H), 7.51 (ddd, J = 8.6, 7.4, 1.6 Hz, 1H), 7.43 (t, J = 7.9 Hz, 2H), 7.22 (td, J = 7.6, 1.2 Hz, 1H), 7.07 (dd, J = 7.5, 1.6 Hz, 1H), 2.28 (tt, J = 11.4, 3.5 Hz, 1H), 1.82 (dd, J = 12.9, 3.5 Hz, 2H), 1.69 (dp, J = 10.6, 3.2 Hz, 2H), 1.60 (dtt, J = 12.7, 3.4, 1.5 Hz, 1H), 1.35 (qd, J = 12.5, 3.2 Hz, 2H), 1.25 (qt, J = 12.6, 3.3 Hz, 2H), 1.14 (qt, J = 12.5, 3.5 Hz, 1H); 19F NMR (376 MHz, DMSO-d6) δ ppm −55.34 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 173.96, 167.09, 140.54, 139.69, 138.49, 137.97, 132.37, 132.08, 131.72, 128.81, 128.29, 128.20, 126.92, 126.73, 126.10, 125.10, 124.27, 123.29, 123.09, 121.78, 120.66, 119.82, 45.20, 29.05, 25.40, 25.10; HRMS (ESI) m/z (M+H) calcd. for C27H26F3N2O2, 467.1941; found 467.1931.
2-(Cyclohexanecarboxamido)-N-(3′-(trifluoromethyl)-[1,1′-biphenyl]-3-yl)benzamide (20)
A mixture of N-(3-bromophenyl)-2-(cyclohexanecarboxamido)benzamide (50.0 mg, 0.125 mmol), 3-(trifluoromethyl)phenylboronic acid (35.5 mg, 0.187 mmol) and Pd(PPh3)4 (7.2 mg, 6.23 μmol) in DMF (l.50 mL) and 2.0 N Na2CO3 (0.50 mL) aqueous solution was heated in μW at 100 °C for 30 min. The reaction was cooled to room temperature, a small portion of Si-THIOL was added to remove palladium. The mixture was filtered and purified via C18 reverse phase HPLC to give the 35 mg (70% yield) of product. LC-MS Retention Time: t1 (Method 1) = 7.438 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.50 (br s, 1H), 10.49 (s, 1H), 8.14–8.24 (m, 1H), 8.01–8.08 (m, 1H), 7.94–8.01 (m, 1H), 7.91 (s, 1H), 7.78–7.84 (m, 2H), 7.70–7.77 (m, 2H), 7.41–7.56 (m, 3H), 7.22 (td, J = 7.6, 1.3 Hz, lH), 2.21–2.35 (m, lH), 1.83 (d, J =l5.8 Hz, 2H), 1.64–1.74 (m, 2H), 1.52–1.63 (m, 1H), 1.08–1.46 (m, 5H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.11 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 173.12, 167.66, 142.23, 139.78, 139.54, 137.09, 135.78, 132.21, 131.29, 129.51, 129.48, 127.76, 127.72, 126.89, 126.84, 124.47, 124.22, 123.65, 122.98, 120.51, 119.86, 43.09, 29.23, 25.11, 24.84; HRMS (ESI) m/z (M+H) calcd. for C27H26F3N2O2, 467.1941; found 467.1943.
2-(Cyclohexanecarboxamido)-N-(3-(trifluoromethyl)benzyl)benzamide (21)
The title compound was prepared according to general protocol A. Yield: 71%; LC-MS Retention Time: t1 (Method 1) = 6.781 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.13 (s, lH), 9.34 (t, J = 6.l Hz, lH), 8.37 (dd, J = 8.4, 1.4 Hz, lH), 7.76 (dd, J = 7.9, 1.7 Hz, lH), 7.66–7.70 (m. lH), 7.58–7.66 (m. 2H), 7.53–7.58 (m, lH), 7.47 (ddd, J = 8.5, 7.2, 1.7 Hz, lH), 7.13 (td, J = 7.6, 1.3 Hz, lH), 4.54 (d, J = 5.9 Hz, 2H), 2.20 (tt, J = l.12, 3.5 lH), 1.75–1.87 (m, 2H), 1.68 (dt, J = 12.3, 3.2 Hz, 2H), 1.53–1.64 (m, lH), 1.02–1.42 (m, 5H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.02 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 173.76, 168.56, 140.60, 139.05, 132.02, 131.43, 129.40, 129.17, 128.86, 128.01, 123.82, 123.78, 123.64, 123.60, 122.54, 120.71, 120.59, 45.69, 42.25, 29.03, 25.35, 25.07; HRMS (ESI) m/z (M+H) calcd. for C22H24F3N2O2, 405.1784; found 405.1790.
2-(Cyclohexanecarboxamido)-N-(furan-2-yl-methyl)benzamide (22)
The title compound was prepared according to general protocol A. Yield: 42%; LC-MS Retention Time: t1 (Method 1) = 6.799 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 12.07 (s, 1H), 10.69 (s, 1H), 9.29 (t, J = 5.8 Hz, 1H), 8.41 (dd, J = 8.3, 1.3 Hz, 1H), 8.22 (dd, J = 8.3, 1.3 Hz, 1H), 7.80 (dd, J = 8.0, 1.6 Hz, 1H), 7.73 (dd, J = 7.8, 1.6 Hz, 1H), 7.45–7.64 (m, 3H), 7.13–7.29 (m, 2H), 6.36 (dd, J = 3.2, 1.9 Hz, 1H), 6.22–6.33 (m, 1H), 4.45 (d, J = 5.5 Hz, 2H), 2.18–2.36 (m, 1H), 1.76–1.91 (m, 2H), 1.64–1.76 (m, 2H), 1.50–1.64 (m, 1H), 1.05–1.44 (m, 5H); 13C NMR (101 MHz, DMSO-d6) δ 174.40, 168.70, 166.89, 152.14, 142.57, 139.06, 138.97, 132.64, 132.60, 128.73, 128.13, 123.82, 122.10, 121.79, 121.73, 110.92, 107.51, 45.81, 36.52, 29.41, 25.82, 25.53; HRMS (ESI) m/z (M+H) calcd. for C26H28N3O4, 446.2074; found 446.2080.
2-Iodo-N-(2-((3-(trifluoromethy1)phenyl)carbamoyl)pheny1)benzamide (24b)
The title compound was prepared according to general protocol C. Yield: 89%; LC-MS Retention Time: t1 (Method 1) = 6.879 min; 1H NMR (400 MHz, DMSO-d6) δ 10.79 (s, 1H), 10.74 (s, 1H), 8.19 – 8.07 (m, 2H), 8.02 – 7.90 (m, 2H), 7.83 (dd, J = 7.8, 1.5 Hz, 1H), 7.67 – 7.41 (m, 5H), 7.34 (td, J = 7.6, 1.2 Hz, 1H), 7.23 (ddd, J = 7.9, 7.1, 2.0 Hz, 1H); 19F NMR (376 MHz, DMSO-d6) δ −61.29 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 167.10, 166.99, 142.26, 139.72, 139.42, 136.99, 131.84, 131.43, 129.83, 129.44, 129.13, 128.93, 128.24, 128.00, 125.64, 124.16, 123.99, 122.75, 122.45, 120.09, 116.59, 116.55, 93.28; HRMS (ESI) m/z (M+H) calcd. for C21H15F3N2O2, 511.0125; found 511.0122.
2-(2-Ethylbutanamido)-N-(3-(trifluoromethyl)phenyl)benzamide (25)
The title compound was pre-pared according to general protocol C. Yield: 12%; LC-MS Retention Time: t1 (Method 1) = 6.751 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.60 (s, lH), 10.25 (s, lH), 8.15 (t, J = 2.2 Hz, lH), 7.98 (dd, J = 8.2, 1.4 Hz, lH), 7.93 (ddd, J =8.2, 1.2, 1.0 Hz, 1H), 7.72 (dd, J = 7.7, 1.7 Hz, 1H), 7.57 (t, J = 7.9 Hz, lH), 7.50 (ddd, J = 8.4, 7.1, 1.7 Hz, lH), 7.38–7.47 (m, lH), 7.23 (td, J = 7.6, 1.3 Hz, 1H), 2.15 (tt, J = 8.8, 5.4 Hz, lH), 1.30–1.62 (m, 4H), 0.74–0.86 (m, 6H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.23 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 176.94, 170.24, 142.91, 140.15, 134.62, 132.92, 132.47, 132.26, 131.68, 129.09, 128.15, 126.93, 126.67, 126.34, 125.82, 123.11, 123.09, 123.06, 119.56, 119.53, 119.50, 53.43, 28.26, 14.82; HRMS (ESI) m/z (M+H) calcd. for C20H22F3N2O2, 379.1628; found 379.1634.
2-Pentanamido-N-(3-(trifluoromethyl)phenyl)benzamide (26)
The title compound was prepared according to general protocol C. Yield: 22%; LC-MS Retention Time: t1 (Method 1) = 6.546 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.61 (s, lH), 10.19 (s, lH), 8.17 (t, J = 2.2 Hz, lH), 7.98 (dd, J =8.l, 1.1 Hz, lH), 7.92 (ddd, J = 8.2, 2.0, 0.8 Hz, lH), 7.71 (dd, J = 7.7, 1.7 Hz, lH), 7.57 (t, J =8.l Hz, 1H), 7.47–7.54 (m, lH), 7.39–7.47 (m, 1H), 7.21 (td, J = 7.6, 1.3 Hz, lH), 2.28 (t, J = 7.4 Hz, 2H), 1.43–1.60 (m, 2H), 1.18–1.34 (m, 2H), 0.80 (t, J = 7.6 Hz, 3H); 19F NMR (376 MHz, DMSO-d6) d ppm −61.21 (s, 3F); 3C NMR (101 MHz, DMSO-d6) δ 171.63, 167.56, 140.24, 137.76, 132.01, 130.23, 129.90, 129.59, 129.08, 125.93, 124.35, 123.79, 122.82, 120.49, 116.95, 116.91, 36.83, 27.59, 22.10, 14.08; HRMS (ESI) m/z (M+H) calcd. for C19H20F3N2O2, 365.1471; found 365.1476.
2-(Cyclopropanecarboxamido)-N-(3-(trifluoromethyl)phenyl)benzamide (27)
The title compound was prepared according to general protocol B. Yield: 51%; LC-MS Retention Time: t1 (Method 1) = 6.131 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.61 (s, lH), 10.46 (s, lH), 8.17 (s, lH), 7.95–8.08 (m, lH), 7.91 (dd, J = 7.9, 1.3 Hz, lH), 7.71 (dd, J = 7.9, 1.5 Hz, lH), 7.57 (t, J = 7.9 Hz, lH), 7.46–7.53 (m, lH), 7.40–7.46 (m, lH), 7.20 (tt, J = 7.6, 0.8 Hz, lH), 1.61–1.81 (m, lH), 0.65–0.79 (m, 4H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.28 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 172.12, 167.57, 140.23, 137.84, 132.08, 130.26, 129.90, 129.13, 125.54, 124.42, 123.69, 122.71, 120.53, 117.02, 116.98, 15.48, 7.85; HRMS (ESI) m/z (M+H) calcd. for C18H16F3N2O2, 349.1158; found 349.1160.
2-(Cyclobutanecarboxamido)-N-(3-(trifiuoromethyl)phenyl)benzamide (28)
The title compound was prepared according to general protocol B. Yield: 53%; LC-MS Retention Time: t1 (Method 1) = 6.412 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.61 (s, lH), 10.22 (s, lH), 8.15 (t, J = 2.2 Hz, lH), 8.09 (dt, J = 8.l, 0.6 Hz, lH), 7.93 (ddd, J = 8.2, 1.2, 1.0 Hz, lH), 7.75 (dd, J = 7.8, 1.6 Hz, 1H), 7.58 (t, J = 7.8 Hz, lH), 7.51 (ddd, J = 8.5, 7.2, 1.6 Hz, 1H), 7.40–7.48 (m, 1H), 7.21 (td, J = 7.6, 1.3 Hz, lH), 3.15–3.26 (m, lH), 2.01–2.24 (m, 4H), 1.80–1.97 (m, 1H), 1.64–1.80 (m, 1H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.18 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 172.88, 167.20, 139.68, 137.72, 131.81, 129.83, 129.45, 128.76, 124.03, 123.15, 121.87, 116.61, 39.73, 24.62, 17.51; HRMS (ESI) m/z (M+H) calcd. for C19H18F3N2O2, 363.1315; found 363.1317.
2-(Cyclopentanecarboxamido)-N-(3-(trifluoromcthyl)phenyl)benzamide (29)
The title compound was prepared according to general protocol B. Yield: 76%; LC-MS Retention Time: t1 (Method 1) = 6.780 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.60 (s, lH), 10.29 (s, lH), 8.15 (t, J = 2.2 Hz, lH), 8.04 (d, J = 7.8 Hz, lH), 7.93 (d, J = 8.4 Hz, lH), 7.73 (dd, J = 7.8, 1.6 Hz, lH), 7.57 (t, J = 7.9 Hz, lH), 7.50 (ddd, J = 8.5, 7.2, 1.6 Hz, lH), 7.44 (d, J = 7.6 Hz, lH), 7.21 (td, J = 7.6, 1.3 Hz, lH), 2.75 (quin, J = 8.0 Hz, lH), 1.74–1.86 (m, 2H), 1.63–1.74 (m, 2H), 1.41–1.63 (m, 4H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.21 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 174.82, 174.17, 167.21, 139.81, 139.79, 137.64, 131.74, 129.88, 129.63, 129.42, 129.21, 129.00, 128.77, 126.90, 125.09, 125.03, 123.98, 123.28, 122.15, 121.48, 120.14, 120.12, 120.09, 120.06, 116.61, 116.59, 116.56, 116.53, 45.67, 29.76, 25.53; HRMS (ESI) m/z (M+H) calcd. for C20H20F3N2O2, 377.1471; found 377.1481.
N-(2-((3(Trifiuoromethyl)phenyl)carbamoyl)phenyl)cycloheptanecarboxamide (30)
The title compound was prepared according to general protocol B. Yield: 70%; LC-MS Retention Time: t1 (Method 1) = 7.093 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.58 (s, lH), 10.18 (s, lH), 8.13 (s, lH), 7.98 (dd, J = 8.l, 0.9 Hz, lH), 7.93 (d, J = 8.0 Hz, lH), 7.71 (dd, J = 7.8, 1.6 Hz, lH), 7.57 (t, J = 7.8 Hz, lH), 7.49 (ddd, J = 8.4, 7.2, 1.6 Hz, lH), 7.39–7.46 (m, lH), 7.21 (td, J =7.6, 1.2 Hz, lH), 2.40–2.51 (m, lH), 1.75–1.88 (m, 2H), 1.31–1.71 (m, lH); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.20 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 174.95, 167.10, 139.75, 137.55, 131.58, 129.80, 129.43, 128.66, 125.33, 123.94, 123.25, 122.31, 120.02, 46.58, 40.15, 27.92, 25.83; HRMS (ESI) m/z (M+H) calcd. for C22H24F3N2O2, 405.1784; found 405.1794.
2-(2-Phenylacetamido)-N-(3-(trifluoromethyl)phenyl)benzamide (31)
The title compound was prepared according to general protocol C. Yield: 85%; LC-MS Retention Time: t1 (Method 1) = 6.443 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.61 (s, lH), 10.30 (s, lH), 8.17 (t, J = 2.l Hz, lH), 8.00–8.07 (m, lH), 7.83–7.91 (m, lH), 7.71 (dd, J = 7.8, 1.6 Hz, lH), 7.57 (t, J = 7.9 Hz, lH), 7.50 (ddd, J = 8.5, 7.2, 1.6 Hz, lH), 7.41–7.47 (m, lH), 7.11–7.34 (m, 6H), 3.66 (s, 2H); 19F NMR (376 MHz, DMSOd6) δ ppm −61.15 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 169.27, 166.93, 139.70, 137.18, 135.28, 131.61, 129.77, 129.45, 129.35, 129.14, 128.55, 128.28, 126.61, 125.52, 125.26, 123.89, 123.51, 122.81, 122.21, 120.09, 116.42, 43.61; HRMS (ESI) m/z (M+H) calcd. for C22H18F3N2O2, 399.1315; found 399.1312.
N-(2-((3-(Trifiuoromethyl)phenyl)carbamoyl)-phenyl)-nicotinamide (32)
The title compound was prepared according to general protocol C. Yield: 55%; LC-MS Retention Time: t1 (Method 1) = 5.342 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.27 (s, lH), 10.74 (s, lH), 8.95–9.12 (m, lH), 8.67–8.91 (m, lH), 8.19–8.29 (m, lH), 8.17 (dd, J = 8.3, 1.3 Hz, lH), 8.10 (t, J = 2.l Hz, lH), 7.92–8.02 (m, lH), 7.86 (dd, J = 7.8, 1.6 Hz, lH), 7.52–7.69 (m, 3H), 7.38–7.48 (m, lH), 7.33 (td, J = 7.6, 1.4 Hz, lH); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.20 (s, 3F); 13C NMR (101 MHz DMSO-d6) δ 167.23, 163.41, 152.11, 148.03, 139.63, 137.31, 135.43, 132.04, 130.28, 129.88, 129.47, 129.16, 129.03, 125.35, 124.19, 123.94, 122.71, 120.21, 116.75, 116.71; HRMS (ESI) m/z (M+H) calcd. for C20H15F3N3O2, 386.1111; found 386.1116.
2-(4-Methoxybenzamido)-N-(3-((trifluoromethyl)phenyl)benzamide (33)
The title compound was prepared according to general protocol C. Yield: 64%; LC-MS RetentionTime: t1 (Method 1) = 6.679 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.31 (s, lH), 10.75 (s, lH), 8.36 (dd, J = 8.3, 1.3 Hz, lH), 8.08 (t, J = 2.2 Hz, lH), 8.01 (ddd, J = 8.2, 1.2, 1.0 Hz, lH), 7.78–7.92 (m, 3H), 7.54–7.64 (m, 2H), 7.42–7.49 (m, lH), 7.27 (td, J = 7.6, 1.2 Hz, lH), 7.00–7.12 (m, 2H), 3.81 (s, 3H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.25 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 167.69, 164.23, 162.19, 139.43, 138.62, 132.31, 129.89, 129.49, 129.17, 129.01, 126.52, 124.48, 123.17, 121.66, 120.41, 117.06, 117.02, 114.07, 55.46, 11.88; HRMS (ESI) m/z (M+H) calcd. for C22H18F3N2O3, 415.1264; found 415.1272.
2-(3-Methoxybenzamido)-N-(3-(trifluoromethyl)phenyl)benzamide (35)
The title compound was prepared according to general protocol C. Yield: 60%; LC-MS Retention Time: t1 (Method 1) = 6.778 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.33 (s, 1H), 10.75 (s, 1H), 8.32 (dd, J = 8.3, 1.3 Hz, 1H), 8.14 (t, J = 2.l Hz, 1H), 7.98 (dt, J = 8.l, 1.2 Hz, 1H), 7.89 (dd, J = 7.9, 1.7 Hz, 1H), 7.52–7.66 (m, 2H), 7.39–7.52 (m, 4H), 7.30 (td, 1.4 Hz, lH), 7.13–7.25 (m, 1H), 3.80 (s, 3H); 19F NMR (376 MHz, DMSOd6) δ ppm −61.31 (s, 3F); HRMS (ESI) m/z (M+H) calcd. for C22H18F3N2O3, 415.1264; found 415.1270.
2-Methoxy-N-(2-((3-(trifluoromethyl)phenyl)carbamoyl)phenyl)benzamide (36)
The title compound was prepared according to general protocol C. Yield: 64%. LC-MS RetentionTime: t1 (Method 1) = 6.734 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.54 (s, lH), 10.84 (s, lH), 8.52–8.63 (m, lH), 8.32 (t, J = 2.2 Hz, lH), 7.94–8.06 (m, 2H), 7.79 (dd, J = 7.7, 1.7 Hz, lH), 7.51–7.68 (m, 3H), 7.43–7.51 (m, lH), 7.24–7.30 (m, lH), 7.17–7.23 (m, lH), 6.99–7.13 (m, lH), 3.97 (s, 3H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.40 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 167.35, 163.02, 157.16, 139.69, 137.66, 133.68, 131.80, 131.52, 129.99, 129.57, 129.26, 128.57, 124.41, 123.72, 123.13, 121.99, 121.32, 120.86, 120.78, 116.30, 116.25, 112.26, 55.59; HRMS (ESI) m/z (M+H) calcd. for C22H13F3N2O3, 415.1264; found 415.1266.
2-Methyl-N-(2-((3-(trifluoromethyl)phenyl)carbamoyl)phenyl)benzamide (37)
The title compound was prepared according to general protocol C. Yield: 46%; LC-MS Retention Time: t1 (Method 1) = 6.769 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.74 (s, 1H), 10.70 (s, 1H), 8.08–8.19 (m, 2H), 7.95 (dt, J = 8.2, 1.2 Hz, lH), 7.79 (dd, J = 7.8, 1.8 Hz, 1H), 7.48–7.65 (m, 3H), 7.33–7.47 (m, 2H), 7.22–7.33 (m, 3H), 2.37 (s, 3H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.30 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 168.70, 167.24, 139.85, 139.02, 137.32, 136.46, 135.88, 131.81, 131.74, 130.91, 130.60, 130.20, 130.10, 129.62, 129.42, 129.21, 129.05, 128.88, 127.06, 125.86, 123.94, 122.57, 116.50, 21.30; HRMS (ESI) m/z (M+H) calcd. for C22H18F3N2O2, 399.1315; found 399.1324.
2-Chloro-N(2-((3-(trifluoromethyl)phenyl)carbamoyl)phenyl)benzamide (38)
The title compound was prepared according to general protocol C. Yield: 81%; LC-MS Retention Time: t1 (Method 1) = 6.765 min; 1H NMR (600 MHz, DMSO-d6) δ 10.82 (s, 1H), 10.71 (s, 1H), 8.12 (d, J = 2.0 Hz, 1H), 8.06 (d, J = 8.2 Hz, 1H), 7.95 – 7.92 (m, 1H), 7.78 (dd, J = 7.8, 1.6 Hz, 1H), 7.62 – 7.51 (m, 3H), 7.48 (td, J = 7.7, 1.7 Hz, 1H), 7.45 – 7.39 (m, 1H), 7.32 (td, J = 7.6, 1.2 Hz, 1H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.28 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 166.97, 164.62, 139.81, 136.68, 136.22, 131.80, 131.58, 130.01, 129.98, 129.89, 129.40, 129.19, 129.03, 128.94, 127.40, 126.26, 125.05, 124.38, 123.93, 122.76, 120.10, 120.07, 116.50, 116.47; HRMS (ESI) m/z (M+H) calcd. for C21H15ClF3N2O2, 419.0769; found 419.0769.
2-(Trifluoromethoxy)-N-(2-((3-(trifluoromethy1)phenyl)carbamoyl)phenyl)benzamide (39)
The title compound was prepared according to general protocol C. Yield: 91%; LC-MS Retention Time: t1 (Method 1) = 6.983 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.97 (s, lH), 10.74 (s, lH), 8.09–8.26 (m, 2H), 7.91–8.03 (m, lH), 7.83 (dd,1=7.8, 1.6 Hz, lH), 7.77 (dd,1=7.6, 2.0 Hz, lH), 7.39–7.70 (m, 6H), 7.33 (td, J = 7.6, 1.3 Hz, lH); 19F NMR (376 MHz, DMSO-d6) δ ppm −56.63 (s, 3F), −61.33 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 166.96, 162.80, 144.79, 139.64, 137.01, 132.53, 131.98, 130.14, 130.00, 129.83, 129.46, 128.90, 127.89, 125.23, 124.15, 124.00, 122.35, 121.83, 120.18, 116.60; HRMS (ESI) m/z (M+H) calcd. for C22H15F6N2O3, 468.0981; found 468.0982.
2-Ethoxy-N(2-((3-(trifluoromethyl)pheny1)carbamoyl)phenyl) benzamide (40)
The title compound was prepared according to general protocol C. Yield: 47%; LC-MS Retention Time: t1 (Method 1) = 6.998 min; 1H NMR (400 MHz, DMSO-d6) δ 11.33 (s, lH), 10.82 (s, lH), 8.51 (dd, J = 8.4, 1.2 Hz, lH), 8.28(t, J = 2.1 Hz, lH), 7.89–8.07 (m, 2H), 7.80 (dd, J = 7.8, 1.6 Hz, lH), 7.55–7.65 (m, 2H), 7.38–7.55 (m, 2H), 7.27 (td, J = 7.6, 1.3 Hz, lH), 7.19 (dd, J = 8.4, 1.0 Hz, lH), 7.01–7.12 (m, lH), 4.30(q, J = 6.9 Hz, 2H), 1.30 (t, J = 6.9 Hz, 3H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.40 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 167.66, 163.81, 156.43, 140.11, 138.08, 133.89, 132.28, 131.95, 130.39, 129.98, 129.66, 129.11, 124.75, 124.19, 123.65, 122.64, 122.57, 121.13, 120.74, 120.70, 116.73, 116.69, 113.48, 64.55, 40.60, 40.40, 40.19, 39.98, 39.77, 39.56, 39.35, 14.27; HRMS (ESI) m/z (M+H) calcd. for C23H20F3N2O3, 429.1421; found 429.1425.
2-Isopropoxy-N-(2-((3-(trifluoromethyl)phenyl)carbamoyl)phenyl)-benzamide (41)
The title compound was prepared according to general protocol H. Yield: 9%; LC-MS Retention Time: t1 (Method 1) = 7.126 min; 1H NMR (600 MHz, DMSO-d6) δ 11.18 (s, 1H), 10.87 (s, 1H), 8.50 (dd, J = 8.3, 1.1 Hz, 1H), 8.36 (t, J = 1.9 Hz, 1H), 7.91 (ddd, J = 11.7, 7.5, 1.7 Hz, 2H), 7.82 (dd, J = 7.8, 1.6 Hz, 1H), 7.63 – 7.57 (m, 2H), 7.54 – 7.44 (m, 2H), 7.29 (td, J = 7.6, 1.2 Hz, 1H), 7.22 – 7.17 (m, 1H), 7.05 (ddd, J = 7.9, 7.3, 1.0 Hz, 1H), 4.82 (p, J = 6.1 Hz, 1H), 1.34 (d, J = 6.0 Hz, 6H); 13C NMR (151 MHz, DMSO-d6) δ 170.14, 166.71, 158.40, 142.76, 140.69, 136.44, 134.96, 134.68, 133.00, 132.55, 132.34, 131.84, 127.27, 126.83, 126.37, 125.83, 125.56, 123.54, 123.37, 123.35, 119.39, 119.36, 117.02, 74.24, 24.10; HRMS (ESI) m/z (M+H) calcd. For C24H22F3N2O3, 443.1577; found 443.1599.
2,6-Dimethoxy-N-(2-((3-(trifluoromethyl)phenyl)carbamoyl)phenyl)benzamide (42)
The title compound was prepared according to general protocol C. Yield: 88%; LC-MS Retention Time: t1 (Method 1) = 6.400 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.68 (s, lH), 10.62 (s, lH), 8.43 (d, J = 8.2 Hz, lH), 8.08 (s, lH), 7.93 (d, J =8.4 Hz, lH), 7.85 (dd, J =7.7, 1.1 Hz, lH), 7.52–7.64 (m, 2H), 7.45 (d, J =7.6 Hz, lH), 7.34 (t, J = 8.4 Hz, lH), 7.27 (td, J = 7.6, 1.1 Hz, lH), 6.71 (d, J = 8.4 Hz, 2H), 3.69 (s, 6H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.29 (s, 3F); 1HRMS (ESI) m/z (M+H) calcd. for C23H20F3N2O4, 445.1370; found 445.1380.
2-(2-Methoxyethoxy)-N-(2-((3-(trifluoromethyl)phenyl)carbamoyl)phenyl)benzamide (43)
The title compound was prepared according to general protocol H. Yield: 16%; LC-MS Retention Time: t1 (Method 1) = 6.625 min; 1H NMR (600 MHz, DMSO-d6) δ 11.25 (s, 1H), 10.84 (s, 1H), 8.45 (dd, J = 8.4, 1.1 Hz, 1H), 8.23 (d, J = 1.9 Hz, 1H), 7.94 (ddd, J = 22.5, 8.1, 2.1 Hz, 2H), 7.82 (dd, J = 7.8, 1.6 Hz, 1H), 7.65 – 7.57 (m, 2H), 7.53 (ddd, J = 8.8, 7.2, 1.8 Hz, 1H), 7.51 – 7.46 (m, 1H), 7.29 (td, J = 7.5, 1.2 Hz, 1H), 7.25 (dd, J = 8.4, 1.0 Hz, 1H), 7.09 (td, J = 7.6, 1.0 Hz, 1H), 4.37 – 4.31 (m, 2H), 3.69 – 3.63 (m, 2H), 3.12 (s, 3H). 19F NMR (376 MHz, DMSO-d6) δ ppm −61.36 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 167.18, 163.39, 156.25, 139.72, 138.97, 137.58, 133.39, 131.90, 131.37, 129.99, 129.49, 129.28, 128.83, 124.55, 123.85, 123.39, 122.40, 122.35, 121.07, 120.29, 116.39, 113.62, 69.79, 67.95, 58.06; HRMS (ESI) m/z (M+H) calcd. for C24H22F3N2O4, 459.1526; found 459.1527.
2-(2-(Dimethylamino)ethoxy)-N-(2-((trifluoromethyl)phenyl)carbamoyl)-phenyl)benzamide (44)
The title compound was prepared according to general protocol H. Yield: 4%; LC-MS Retention Time: t1 (Method 1) = 4.705 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.11 (s, 1H), 10.84 (s, 1H), 8.41 (dd, J = 8.5, 1.3 Hz, 1H), 8.07–8.20 (m, 1H), 7.96 (dd. J =8.0. 1.2 Hz, 1H), 7.87 (dd, J = 7.7. 1.9 Hz, 1H), 7.84 (dd, J = 7.8, 1.8 Hz, 1H). 7.57–7.65 (m, 2H), 7.52–7.57 (m, 1H), 7.45–7.52 (m, 1H), 7.24–7.33 (m, 2H), 7.13 (td, J = 7.5, 1.0 Hz, 1H), 4.55 (t, J = 5.l Hz, 2H), 3.49–3.74 (m, 2H), 2.78 (s, 6H); HRMS (ESI) m/z (M+H) calcd. for C25H25F3N3O3, 472.1843; found 472.1851.
2-(2-Ethoxybenzamido)-N-(3-((trifluoromethyl)sulfonyl)phenyl)nicotinamide (49)
The title compound was prepared according to general protocol A. Yield: 45%; LC-MS Retention Time: t1 (Method 1) = 5.472 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.98 (s, lH), 10.81 (s, lH), 8.47–8.66 (m, 2H), 8.05–8.24 (m, 2H), 7.75–7.87 (m, 2H), 7.72 (dd, J = 7.8, 2.0 Hz, lH), 7.51 (ddd, J = 8.5, 7.1, 2.0 Hz, lH), 7.39 (dd, J = 7.6, 4.9Hz, lH), 7.20 (d, J = 8.2Hz, lH), 7.00 (t, J = 7.4 Hz, lH), 4.27 (q, J = 7.0 Hz, 2H), 1.42 (t, J =7.0 Hz, 3H); 19F NMR (376 MHz, DMSO-d6) δ ppm −78.44 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 165.91, 163.44, 156.44, 150.26, 148.04, 141.24, 137.87, 133.55, 131.38, 130.94, 129.86, 128.00, 125.15, 124.02, 121.63, 120.76, 120.51, 120.43, 120.27, 118.35, 113.33, 64.71, 14.44; HRMS (ESI) m/z (M+H) calcd. for C22H19F3N3O5S, 494.0992; found 494.1004.
3-(2-Ethoxybenzamido)-N-(3-((trifluoromethyl)sulfonyl)phenyl)isonicotinamide (50)
The title compound was prepared according to general protocol A. Yield: 38%; LC-MS Retention Time: t1 (Method 1) = 5.766 min; HRMS (ESI) m/z (M+H) calcd. for C22H19F3N3O5S, 494.0992; found 494.1000.
4-(2-Ethoxybenzamido)-N-(3-((trifluoromethyl)sulfonyl)phenyl)nicotinamide (51)
The title compound was prepared according to general protocol A. Yield: 26%; LC-MS Retention Time: t1 (Method 1) = 5.507 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.71 (s, lH), 11.25 (s, lH), 9.00 (s, lH), 8.77 (t, J = 2.2 Hz, lH), 8.65–8.71 (m, lH), 8.54–8.64 (m, lH), 8.15–8.26 (m, lH), 7.96 (dd, .J = 7.8, 2.3 Hz, lH), 7.78–7.93 (m, 2H), 7.56 (ddd, J =8.6, 7.0, 2.0 Hz, lH), 7.24 (d, J = 8.6 Hz, lH), 7.09 (t, J =7.4 Hz, lH), 4.35 (q, J = 6.9 Hz, 2H), 1.29 (t, J = 6.8 Hz, 3H); 19F NMR (376 MHz, DMSO-d6) δ ppm −78.37 (s, 3F); 13C NMR (151 MHz, DMSO-d6) δ 169.07, 167.39, 159.21, 156.20, 153.04, 147.91, 143.56, 137.33, 134.89, 134.57, 133.02, 131.82, 129.06, 124.40, 124.05, 123.94, 123.60, 121.44, 120.83, 117.83, 116.25, 67.26, 16.85; HRMS (ESI) m/z (M+H) calcd. for C22H19F3N3O5S, 494.0992; found 494.1004.
2-(2-Ethoxybenzamido)-N-(3-((trifluoromethyl)sulfonyl)phenyl)-nicotinamide (52)
The title compound was prepared according to general protocol A. yield: 51%; LC-MS Retention Time: t1 (Method 1) = 7.402 min; HRMS (ESI) m/z (M+Na)+ calcd. for C22H18F3N3NaO5S, 516.0811; found 516.0837.
2-(2-Ethoxyphenylsulfonamido)-N-(3-((trifluoromcthy1)sulfonyl)phenyl)-benzamide (53)
The title compound was prepared according to general protocol A. Yield: 29%; LC-MS Retention Time: t1 (Method 1) = 6.631 min; 1H NMR (400 MHz, DMSO-d6) δ 11.00 (s, 1H), 10.37 (s, 1H), 8.64 (t, J = 1.9 Hz, 1H), 8.24 (dt, J = 7.8, 1.8 Hz, 1H), 7.96 – 7.84 (m, 2H), 7.82 (dd, J = 7.8, 1.7 Hz, 2H), 7.60 – 7.50 (m, 2H), 7.48 (ddd, J = 8.5, 7.3, 1.5 Hz, 1H), 7.18 (td, J = 7.6, 1.2 Hz, 1H), 7.12 – 7.00 (m, 2H), 3.95 (q, J = 7.0 Hz, 2H), , 1.18 (t, J = 7.0, 3H); 19F NMR (376 MHz, DMSO-d6) δ −78.37; 13C NMR (101 MHz, DMSO-d6) δ 167.47, 155.62, 140.29, 137.78, 135.61, 133.03, 131.39, 130.49, 129.95, 129.46, 128.91, 125.92, 125.37, 123.06, 121.39, 121.31, 121.05, 120.03, 118.75, 117.80, 113.56, 64.53, 13.68; HRMS (ESI) m/z (M+H) calcd. for C22H20F3N2O6S2, 529.0709; found 529.0716.
2-Methoxy-N-methy1-N-(2-((3-(trifluoromethyl)pheny1)carbamoy1)phenyl)benzamide (54)
The title compound was prepared according to general protocol A. Yield: 47%; LC-MS Retention Time: t1 (Method 1) = 6.046 min; HRMS (ESI) m/z (M+H) calcd. for C23H20F3N2O3, 429.1421; found 429.1438.
2-(2-Methoxybenzamido)-N-methyl-N-(3-(trifluoromethyl)phenyl) benzamide (55)
The title compound was prepared according to general protocol A. Yield: 60%; LC-MS Retention Time: t1 (Method 1) = 6.442 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.65 (d, J = 0.8 Hz, lH), 8.21 (dt, J = 8.4, 0.6 Hz, lH), 8.03 (dd, J = 7.8, 1.8 Hz, lH), 7.54–7.66 (m, 2H), 7.37–7.54 (m, 3H), 7.20–7.33 (m, 2H), 7.13 (ddd, J = 7.9, 7.2, 1.0 Hz, lH), 7.04–7.11 (m, lH), 6.86–6.96 (m, lH), 4.02 (s, 3H), 3.44 (s, 3H); HRMS (ESI) m/z (M+H) calcd. for C23H20F3N2O3, 429.1241; found 429.1418.
2-((2-Methoxyethyl)amino)-N-(3-(trifiuoromethyl)phenyl)-benzamide (57)
The title compound was prepared according to general protocol F. Yield: 19%; LC-MS Retention Time: t1 (Method 1) = 6.254 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.33 (s, lH), 8.14 (t, J = 2.2 Hz, lH), 7.88–8.05 (m, lH), 7.70 (dd, J = 7.8, 1.6 Hz, lH), 7.54–7.63 (m, lH), 7.44–7.52 (m, lH), 7.37–7.44 (m, lH), 7.34 (ddd, J = 8.6, 7.0, 1.7 Hz, lH), 6.72–6.84 (m, lH), 6.65 (ddd, J = 8.0, 7.1, 1.2 Hz, lH), 3.53 (t, J = 5.5 Hz, 2H), 3.28 (s, 3H), 3.24–3.33 (m, 2H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.23 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 168.36, 149.33, 139.94, 133.15, 129.75, 129.19, 129.08, 124.02, 119.73, 116.55, 116.51, 114.97, 114.37, 111.30, 70.34, 58.03, 41.90; HRMS (ESI) m/z (M+H) calcd. for C17H18F3NO2, 339.1315; found 339.1326.
2-(2-Ethoxybenzamido)-N-(3-((trifluoromethyl)sulfonyl)phenyl)thiophene-3-carboxamide (59)
The title compound was prepared according to general protocol A. Yield: 33%; LC-MS Retention Time: t1 (Method 1) = 7.341 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 13.25 (s, 1H), 10.45 (s, 1H), 8.90 (d, J = 2.2 Hz, 1H), 8.30 – 8.20 (m, 1H), 8.08 (dd, J = 7.9, 1.9 Hz, 1H), 7.90– 7.80 (m, 2H), 7.75 (d, J = 6.0 Hz, 1H), 7.60 (ddd, J = 8.9, 7.3, 1.9 Hz, 1H), 7.31 (dd, J = 8.6, 0.9 Hz, 1H), 7.18–7.08 (m, 2H), 4.51 (q, J = 7.0 Hz, 2H), 1.47 (t, J = 7.0 Hz, 3H). 13C NMR (101 MHz, DMSO-d6) δ 164.07, 162.19, 156.99, 147.77, 141.20, 134.92, 132.43, 131.69, 130.33, 129.06, 125.79, 123.30, 121.47, 121.39, 119.79, 117.58, 115.83, 113.79, 64.98, 14.46; 19F NMR (376 MHz, DMSO-d6) δ −78.43 (s, 3F); HRMS (ESI) m/z (M+H) calcd. for C21H18F3N2O5S2, 499.0604; found 499.0608.
2-Methoxy-N-(3-((3-(trifluoromethyl)phenyl)carbamoyl)phenyl)benzamide (61)
The title compound was prepared according to general protocol A. Yield: 68%; LC-MS Retention Time: t1 (Method 1) = 6.471 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 10.58 (s, lH), 10.30 (s, lH), 8.27–8.35 (m, lH), 8.23 (t, J = 2.l Hz, lH), 8.04 (dt, J =8.2, 1.1 Hz, lH), 7.88–7.99 (m, lH), 7.67 (ddd, J = 8.0, 1.4, 1.2 Hz, lH), 7.63 (dd, J = 7.6, 1.8 Hz, lH), 7.56–7.62 (m, lH), 7.47–7.56 (m, 2H), 7.41–7.47 (m, lH), 7.14–7.23 (m, lH), 7.06 (td, J = 7.4, l.0 Hz, lH), 3.90 (s, 3H); 19F NMR (376 MHz, DMSO-d6) δ ppm −61.28 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 166.43, 165.23, 156.95, 140.40, 139.69, 135.69, 132.59, 130.32, 130.08, 129.95, 129.24, 125.95, 125.21, 124.18, 123.45, 122.99, 120.93, 119.66, 116.76, 116.72, 112.46; HRMS (ESI) m/z (M+H) calcd. for C22H18F3N2O3, 415.1264; found 415.1275.
2-Methoxy-N-(2-((3-((trifluoromethyl)sulfonyl)phenyl)carbamoy1)phenyl)benzamide (62)
The title compound was prepared according to general protocol G. Yield: 25%; LC-MS Retention Time: t1 (Method 1) = 6.810 min; 1H NMR (400 MHz, DMSO-d6) δ ppm 11.58 (s, 1H), 11.09 (s, 1H), 8.82 (t, J = 2.l Hz, 1H), 8.58 (dd,1=8.5, 1.3 Hz, lH), 8.23 (dt, J = 7.0, 2.2 Hz, lH), 8.02 (dd, J = 7.7, 1.9 Hz, lH), 7.76–7.99 (m, 3H), 7.51–7.63 (m, 2H), 7.27 (td, J = 7.5, 1.2 Hz, 1H), 7.22 (dd, J = 8.4, 1.2 Hz, 1H), 7.09 (ddd, J = 7.2, 1.2 Hz, lH), 3.99 (s, 3H); 19F NMR (376 MHz, DMSO-d6) δ ppm −78.39 (s, 3F); 13C NMR (101 MHz, DMSO-d6) δ 167.57, 163.01, 157.16, 140.75, 137.78, 133.69, 132.05, 131.51, 131.37, 129.94, 128.69, 128.43, 125.58, 123.90, 123.10, 122.05, 121.24, 120.84, 120.77, 112.26, 55.59; HRMS (ESI) m/z (M+H) calcd. for C22H18F3N2O5S, 479.0883; found 479.0886.
2-Ethoxy-N-(2-((3-((trifluoromethyl)sulfonyl)phenyl)carbamoyl)phenyl)benzamide (63)
The title compound was prepared according to general protocol A. Yield: 53%; LC-MS Retention Time: t1 (Method 1) = 6.963 min; 1H NMR (400 MHz, DMSO-d6): δ 11.35 (s, 1H), 11.10 (s, 1H), 8.76 (t, J = 2.2 Hz, 1H), 8.53 (d, J = 8.2 Hz, 1H), 8.23 (dt, J = 7.1, 2.3 Hz, 1H), 7.96 (dd, J = 7.8, 2.0 Hz, 1H), 7.80–7.92 (m, 3H), 7.57 – 7.65 (m, 1H), 7.53 (ddd, J = 8.6, 7.0, 2.0 Hz, 1H), 7.30 (td, J = 7.6, 1.2 Hz, 1H), 7.22 (d, J = 8.2 Hz, 1H), 7.04–7.10 (m, 1H), 4.31 (q, J = 6.8 Hz, 2H), 1.31 (t, J = 6.8 Hz, 3H); 13C NMR (101 MHz, DMSO-d6): δ 167.44, 163.39, 156.00, 140.75, 137.73, 133.44, 132.10, 131.48, 131.34, 129.93, 128.81, 128.49, 125.58, 123.86, 123.21, 122.27, 122.08, 120.81, 120.67, 119.41 (q, J = 328 Hz), 113.00, 64.09, 13.80; 19F NMR (376 MHz, DMSO-d6): δ −78.40 (s, 3F); HRMS (m/z): [M+H] calcd. for C23H20F3N2O5S, 493.1040; found 493.1044.
2-Isopropoxy-N-(2-(3-(trifluoromethylsulfonyl)phenylcarbamoyl)phenyl)benzamide (64)
The title compound was prepared according to general protocol A. Yield: 44%; LC-MS Retention Time: t1 (Method 1) = 6.804 min; 1H NMR (400 MHz, DMSO-d6): δ 11.10 (s, 1H), 11.05 (s, 1H), 8.64 (s, 1H), 8.44 (d, J = 8.2 Hz, 1H), 8.27 (dt, J = 7.1, 2.3 Hz, 1H), 7.79–7.92 (m, 4H), 7.60–7.69 (m, 1H), 7.52–7.60 (m, 1H), 7.37 (d, J = 8.2 Hz, 1H), 7.31 (t, J = 7.6 Hz, 1H), 7.19 (t, J = 7.4 Hz, 1H), 4.94 (q, J = 9.0 Hz, 2H); 13C NMR (101 MHz, DMSO-d6): δ 167.46, 163.36, 154.24, 140.76, 137.68, 132.87, 132.27, 131.25, 130.85, 129.87, 128.99, 128.60, 125.45, 124.28, 123.83, 123.63 (q, J = 280 Hz), 123.49, 122.52, 121.97, 121.05, 119.42 (q, J = 329 Hz), 114.03, 65.00 (q, J = 35.0 Hz); 19F NMR (376 MHz, DMSOd6): δ 72.46 (t, J = 9.4 Hz, 3F), −78.48 (s, 3F); HRMS (m/z): [M+H] calcd. for C23H17F6N2O5S, 547.0757; found 547.0762.
2-Isopropoxy-N-(2-(3-(trifluoromethylsulfonyl)phenylcarbamoyl)phenyl)benzamide (65)
The title compound was prepared according to general protocol A. Yield: 57%; LC-MS Retention Time: t1 (Method 1) = 6.974 min; t2 (Method 2) = 3.881 min; 1H NMR (400 MHz, DMSO-d6): δ 11.11 (s, 1H), 11.09 (s, 1H), 8.75 (s, 1H), 8.48 (d, J = 8.2 Hz, 1H), 8.20 (dt, J = 7.4, 2.2 Hz, 1H), 7.91 (dd, J = 7.8, 1.6 Hz, 1H), 7.77–7.89 (m, 3H), 7.57–7.66 (m, 1H), 7.51 (ddd, J = 8.6, 7.0, 2.0 Hz, 1H), 7.30 (td, J = 7.5, 1.0 Hz, 1H), 7.20 (d, J = 8.6 Hz, 1H), 7.05 (t, J = 7.4 Hz, 1H), 4.75 – 4.85 (m, 1H), 1.32 (d, J = 5.9 Hz, 6H); 13C NMR (101 MHz, DMSO-d6): δ 167.24, 163.63, 155.29, 140.75, 137.61, 133.30, 132.06, 131.52, 131.28, 129.97 (br.), 128.89, 128.51, 125.58, 123.93, 123.29, 122.79, 122.56, 120.82, 120.44, 119.40 (q, J = 328 Hz), 113.98, 71.19, 21.05; 19F NMR (376 MHz, DMSO-d6): δ −78.38 (s, 3F); HRMS (m/z): [M+H] calcd. for C24H22F3N2O5S, 507.1196; found 507.1221.
2-Propoxy-N-(2-((3-((trifluoromethyl)sulfonyl)phenyl)carbamoyl)phenyl)benzamide (66)
The title compound was prepared according to general protocol A. Yield: 41%; LC-MS Retention Time: t1 (Method 1) = 7.147 min; 1H NMR (400 MHz, DMSO-d6): δ 11.26 (s, 1H), 11.10 (s, 1H), 8.72 (s, 1H), 8.54 (d, J = 8.6 Hz, 1H), 8.24 (dt, J = 7.1, 2.3 Hz, 1H), 7.94 (dd, J = 7.8, 2.0 Hz, 1H), 7.78 – 7.91 (m, 3H), 7.56 – 7.67 (m, 1H), 7.52 (ddd, J = 8.5, 7.1, 2.0 Hz, 1H), 7.25–7.33 (m, 1H), 7.20 (d, J = 8.2 Hz, 1H), 7.07 (t, J = 7.4 Hz, 1H), 4.16 (t, J = 7.0 Hz, 2H), 1.71 (sxt, J = 7.2 Hz, 2H), 0.77 (t, J = 7.4 Hz, 3H); 13C NMR (101 MHz, DMSO-d6): δ 167.42, 163.49, 156.14, 140.78, 137.75, 133.37, 132.14, 131.39, 131.33, 129.95 (br.), 128.90, 128.48, 125.54, 123.80, 123.21, 122.25, 122.15, 120.88, 120.68, 119.43 (q, J = 328 Hz), 113.09, 69.82, 21.24, 9.74; 19F NMR (376 MHz, DMSO-d6): δ −78.43 (s, 3F); HRMS (m/z): [M+H] calcd. for C24H22F3N2O5S, 507.1196; found 507.1219.
2-Butoxy-N-(2-((3-((trifluoromethyl)sulfonyl)phenyl)carbamoyl)phenyl)benzamide (67)
The title compound was prepared according to general protocol A. Yield: 49%; LC-MS Retention Time: t1 (Method 1) = 7.325 min; t2 (Method 2) = 3.971 min; 1H NMR (400 MHz, DMSO-d6): δ 11.27 (s, 1H), 11.10 (s, 1H), 8.71 (s, 1H), 8.55 (d, J = 8.2 Hz, 1H), 8.26 (dt, J = 7.3, 2.0 Hz, 1H), 7.93 (dd, J = 7.8, 2.0 Hz, 1H), 7.81–7.91 (m, 3H), 7.58–7.68 (m, 1H), 7.47–7.57 (m, 1H), 7.29 (t, J = 7.4 Hz, 1H), 7.22 (d, J = 8.2 Hz, 1H), 7.08 (t, J = 7.4 Hz, 1H), 4.19 (t, J = 6.8 Hz, 2H), 1.57–1.73 (m, 2H), 1.23 (sxt, J = 7.5 Hz, 2H), 0.70 (t, J = 7.4 Hz, 3H); 13C NMR (101 MHz, DMSO-d6): δ 167.41, 163.52, 156.13, 140.81, 137.81, 133.37, 132.20, 131.36, 131.34, 129.96 (br.), 128.94, 128.41, 125.54, 123.59, 123.18, 122.30, 122.06, 120.85, 120.69, 113.09, 68.19, 29.93, 18.34, 13.37; 19F NMR (376 MHz, DMSO-d6): δ −78.42 (s, 3F); HRMS (m/z): [M+H] calcd. for C25H24F3N2O5S, 521.1353; found 521.1359.
Supplementary Material
Highlights.
A high throughput screening campaign identified the first small molecule relaxin peptide hormone receptor agonists
An extensive medicinal chemistry effort produced an optimized, potent lead compound
Pharmokinetic studies show good compound bioavailability in mice with no observed toxicity
The lead compound displays an antifibrotic gene expression profile in activated human hepatic stellate cells
Acknowledgments
This research was supported by the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research Grant U54MH084681 and the Intramural Research Program of the National Center for Advancing Translational Sciences at the National Institutes of Health. Dr. Daniel C. Talley and Dr. Scott B. Hoyt and are acknowledged for critical editing of this manuscript. The authors thank Dr. Christopher LeClair and Emmett George for analytical and technical assistance.
ABBREVIATIONS USED
- AcOH
acetic acid
- aq
aqueous
- CH2Cl2
dichloromethane
- conc
concentrated
- DIPEA
diisopropylethylamine
- DMAP
dimethylaminopyridine
- DME
1,2-dimethoxyethane
- DMF
N,Ndimethylformamide
- DMSO
dimethyl sulfoxide
- EDC
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide
- Et2O
diethyl ether
- EtOAc
ethyl acetate
- EtOH
ethanol
- H2O
water
- HATU
1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxide hexafluorophosphate
- HCl
hydrochloric acid
- iPr2NH
diisopropylamine
- IV
intervenous
- K2CO3
potassium carbonate
- KOAc
potassium acetate
- LCMS
liquid chromatography/mass spectrometry
- M
molar
- mM
millimolar
- MeOH
methanol
- MHz
megahertz
- N2
nitrogen
- NaHCO3
sodium bicarbonate
- Na2CO3
sodium carbonate
- NaOH
sodium hydroxide
- NH4Cl
ammonium chloride
- PBS
phosphate-buffered saline
- Pd(dppf)Cl2·CH2Cl2
[1,1]-bis(diphenylphosphino)ferrocene]dichloropalladium (II) complex with dichloromethane
- Pd(PPh3)4
tetrakis(triphenylphosphine)palladium (0)
- PO
by mouth
- RXFP
relaxin/insulin-like family peptide receptor
- TFA
trifluoroacetic acid
- THF
tetrahydrofuran
- TGF
tumor growth factor
- TNF
tumor necrosis factor
- μW
microwave irradiation.
Footnotes
Mouse PK data, crystal structure of compound 65 and X-ray data, carbon, proton and fluorine NMR spectra of selected compounds including compound 65, maximum tolerated dose study for compound 65
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hisaw FL. Experimental relaxation of the pubic ligament of the guinea pig. Proc Soc Exp Biol Med. 1926;23(8):661–663. [Google Scholar]
- 2.Fevold HL, Hisaw FL, Meyer RK. The relaxative hormone of the corpus luteum. Its purification and concentration. J Am Chem Soc. 1930;52(8):3340–3348. [Google Scholar]
- 3.Conrad KP. Maternal vasodilation in pregnancy: the emerging role of relaxin. Am J Physiol. 2011;301(2):R267–R275. doi: 10.1152/ajpregu.00156.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeyabalan A, Shroff SG, Novak J, Conrad KP. The vascular actions of relaxin. In: Agoulnik AI, editor. Relaxin and related peptides. Vol. 612. Springer; New York: 2007. pp. 65–87. [DOI] [PubMed] [Google Scholar]
- 5.Schrier RW, Durr JA. Pregnancy: an overfill or underfill state. Am J Kidney Dis. 1987;9:284–289. doi: 10.1016/s0272-6386(87)80123-3. [DOI] [PubMed] [Google Scholar]
- 6.Conrad KP, Shroff SG. Effects of relaxin on arterial dilation, remodeling, and mechanical properties. Curr Hypertens Rep. 2011;13:409–420. doi: 10.1007/s11906-011-0231-x. [DOI] [PubMed] [Google Scholar]
- 7.Teichman SL, Unemori E, Teerlink JR, Cotter G, Metra M. Relaxin: Review of biology and potential role in treating heart failure. Curr Heart Failure Rep. 2010;7:75–82. doi: 10.1007/s11897-010-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Der Westhuizen ET, Summers RJ, Halls ML, Bathgate RAD, Sexton PM. Relaxin receptors – New drug targets for multiple disease states. Current Drug Targets. 2007;8:91–104. doi: 10.2174/138945007779315650. [DOI] [PubMed] [Google Scholar]
- 9.Patil NA, Rosengren KJ, Separovic F, Wade JD, Bathgate RAD, Hossain A. Relax-in family peptides: structure – activity relationship studies. Br J Pharmacol. 2017;174(10):950–961. doi: 10.1111/bph.13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong RCK, Petrie EJ, Mohanty B, Ling J, Lee JC, Gooley PR, Bathgate RAD. The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation. J Biol Chem. 2013;288(39):28138–28151. doi: 10.1074/jbc.M113.499640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dessauer CW, Nguyen BT. Relaxin stimulates multiple signalingpathways. Ann NY Acad Sci. 2005;1041:272–279. doi: 10.1196/annals.1282.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halls ML, Bathgate RAD, Summers RJ. Comparison of signaling pathways activiared by the relaxin family peptide receptors, RXFP1 and RXFP2, using reporter genes. J Pharmacol Exp Ther. 2007;320(1):281–290. doi: 10.1124/jpet.106.113225. [DOI] [PubMed] [Google Scholar]
- 13.Ryan PL, Youngblood RC, Harvill J, Willard ST. Photonic monitoring in real time of vascular endothelial growth factor receptor 2 gene expression under relaxin-induced conditions in a novel murine wound model. Ann N Y Acad Sci. 2005;1041:398–414. doi: 10.1196/annals.1282.061. [DOI] [PubMed] [Google Scholar]
- 14.Teichman SL, Unemori E, Dschietzig T, Conrad K, Voors AA, Teerlink JR, Felker GM, Cotter G, Metra M. Relaxin, a pleiotropic vasodilator for the treatment of heart failure. Heart Fail Rev. 2009;14:321–329. doi: 10.1007/s10741-008-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teerlink JR, Voors AA, Ponikowski P, pang PS, greenberg BH, Filippatos G, Felker GM, Davison BA, Cotter G, Gimpelewicz C, Boer-Martins L, Wernsing M, Hua TA, Severin T, Metra M. Serelaxin in addition to standard therapy in acute heart failure: rationale and design of the RELAX-AHF-2 study. Eur J Heart Fail. 2017;19:800–809. doi: 10.1002/ejhf.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metra M, Shoaib A, Mabote T, Zuhair M, Kassianides X, Cleland JGF. Effect of serelaxin on cardiac renal and hepatic biomarkers in the relaxin in acute heart failure (RELAX-AHF) development program: Correlation with outcomes. J Am Coll Cardiol. 2013;61:196–206. doi: 10.1016/j.jacc.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Xiao J, Huang Z, Chen CZ, Agoulnik IU, Southall N, Hu X, Jones RE, Ferrer M, Zheng W, Agoulnik AI, Marugan JJ. Identification and optimization of small-molecule agonists of the human relaxin hormone receptor RXFP1. Nat Commun. 2013;4:1953. doi: 10.1038/ncomms2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Myhr C, Huang Z, Xiao J, Barnaeva E, Ho BA, Agoulnik IU, Ferrer M, Marugan JJ, Southall N, Agoulnik AI. Structural insights into the activation of human relaxin family peptide receptor 1 by small-molecule agonists. Biochemistry. 2016;55:1772–1783. doi: 10.1021/acs.biochem.5b01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocan M, Sarwar M, Ang SY, Xiao J, Marugan JJ, Hossain MA, Wang C, Hutchinson DS, Samuel CS, Agoulnik AI, Bathgate RAD, Summer RJ. ML290 is a biased allosteric agonist at the relaxin receptor RXFP1. Nat Commun. 2017;7:2968. doi: 10.1038/s41598-017-02916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.For all biological assays, materials and methods see reference 17.
- 21.Dandapani S, Marcaulle LA. Accessing new chemical space for ‘undruggable’ targets. Nat Chem Biol. 2010;6:861. doi: 10.1038/nchembio.479. [DOI] [PubMed] [Google Scholar]
- 22.Halls ML, van der Westhuizen ET, Bathgate RA, Summers RJ. Relaxin family peptide receptors: former orphans reunite with their parent ligands to activate multiple signaling pathways. Br J Pharmacol. 2007;150:677. doi: 10.1038/sj.bjp.0707140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalgutkar AS, Gardner I, Obach RS, Shaffer CL, Callegari E, Henne KR, Mutlib AE, Dalvie DK, Lee JS, Nakai Y, O’Donnell JP, Boer J, Harriman SP. Curr Drug Metab. 2005;6:197. doi: 10.2174/1389200054021799. [DOI] [PubMed] [Google Scholar]
- 24.Samuel CS, Royce SG, Hewitson TD, Denton KM, Cooney TE, Bennett RG. Anti-fibrotic actions of relaxin. Br J Pharmacol. 2016;174:962. doi: 10.1111/bph.13529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett RG, Dalton SR, Mahan KJ, Gentry-Nielsen MJ, Hamel FG, Tuma DJ. Relaxin receptors in hepatic stellate cells and cirrhotic liver. Biochem Pharmacol. 2007;73:1033. doi: 10.1016/j.bcp.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Fallowfield JA, Hayden AI, Snowden VK, Aucott RL, Stuchfield BM, Mole DJ, Pellicoro A, Gordon-Walker TT, Henke A, Schrader J, Trivedi PJ, Princivalle M, Forbes SJ, Collins JE, Iredale JP. Relaxin modulates human and rat hepatic myofibroblast function and ameliorates portal hypertension in vivo. Hepatology. 2014;59:1492. doi: 10.1002/hep.26627. [DOI] [PubMed] [Google Scholar]
- 27.Singh S, Bennett RG. Relaxin signaling activates peroxisome proliferator-activated receptor gamma. Mol Cell Endocrinol. 2010;315:239. doi: 10.1016/j.mce.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leeuw A, De M, Mccarthy SP, Geerts A, Knook DL. Purified rat liver fat-storing cells in culture divide and contain collagen. Hepatology. 1984;4:392. doi: 10.1002/hep.1840040307. [DOI] [PubMed] [Google Scholar]
- 29.McBride A, Hoy AM, Bamford MJ, Mossakowska DE, Ruediger MP, Griggs J, Desai S, Simpson K, Caballero-Hernandez I, Iredale JP, Pell T, Aucott RL, Holmes DS, Webster SH, Fallowfield JA. In search of a small molecule agonist of the relaxin receptor RXFP1 for the treatment of liver fibrosis. Sci Rep. 2017;7:10806. doi: 10.1038/s41598-017-10521-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kern A, Hubbard D, Amano A, Bryant-Greenwood GD. Cloning, expression and functional characterization of relaxin receptor (leucine-rich repeat-containing g protein-coupled receptor 7) splice variants from human fetal membranes. Endocrinology. 2008;149:1277. doi: 10.1210/en.2007-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu SY, et al. Activation of orphan receptors by the hormone relaxin. Science. 2002;295:671. doi: 10.1126/science.1065654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.