Abstract
Here, we describe a novel method for fabrication of microfluidic structures in classroom environments. This method is based on replica moulding of pasta structures in polydimethylsiloxane. Placing pasta structures on a petroleum jelly base layer enables templating round-shaped structures with controllable cross-sectional profiles. The pasta structures can be easily deformed and combined to create more complex 3D microfluidic structures. Proof-of-concept experiments indicate the capability of this method for studying the mixing of neighbouring flows, generation of droplets, lateral migration of particles, as well as culturing, shear stress stimulation, and imaging of cells. Our “do-it-in-classroom” method bridges the gap between the classroom and the laboratory.
INTRODUCTION
Microfluidics involves the manipulation of fluids in microscale structures. It offers portable, disposable, and inexpensive lab-on-a-chip devices for various biomedical applications such as point-of-care diagnostics,1 immunoassays,2,3 drug delivery,4 and various cellular studies.5 The last decade has experienced a consistent increase in companies and commercially available products taking advantage of microfluidic technologies.6 Further advancement of microfluidics technologies relies on training skilled students and researchers at universities. This requires the effective inclusion of hands-on activities, laboratories, and mini-projects in the current lecture-based courses to provide students with both theoretical and practical skills required to develop microfluidic systems. More importantly, the multi-disciplinary nature of microfluidics requires these courses to be suitable for students with various engineering, science, and biomedical backgrounds. Given the development of a microfluidic device involves design, fabrication, and characterisation steps, the success of such an interactive microfluidic course depends on the successful inclusion of the “fabrication step” in the curriculum of the course.
There are four commonly used fabrication techniques in microfluidics: photolithography, injection moulding, laser cutting, and 3D printing. Photolithography involves creating a master for templating micro/nano scale structures7 and is suitable for parallel fabrication of 2D structures.8 It can also be used for the fabrication of 3D structures, although tedious alignment processes might be required.9 Importantly, photolithography requires multi-million-dollar cleanroom facilities and microfabrication expertise.7 Injection moulding, on the other hand, involves injecting molten thermoplastic granules into a pre-made mould.10–12 It enables mass production of microstructures with minimal labour to realise 3D multilayer or high aspect ratio structures.13 Laser cutting involves the removal of a material via a high temperature laser beam (e.g., CO2 laser) to directly fabricate microfluidic chips in a one-step process.14,15 It is suitable for rapid prototyping, although the laser can cause undesired rough surfaces.14 3D printing involves making layers of a material to fabricate the desired structure.16,17 It facilitates the fabrication of 3D structures with complex geometries in a relatively quick process,18–20 thus it is suitable for rapid prototyping using a wide variety of materials.21–24
The majority of universities do not have microfabrication facilities. Even when universities have such facilities (for example, here at RMIT University, we have access to photolithography, laser cutting, and 3D printing technologies), the amount of time and resources required for training students within a 12-week period can still limit the effective inclusion of fabrication into the microfluidics course. Hence, the adoption of “do-it-in-classroom” fabrication approaches can potentially benefit the running of such interactive microfluidic courses. These approaches should enable the fabrication of microfluidic structures: (i) in the classroom environment without the need for microfabrication facilities and (ii) without necessarily having background knowledge in CAD software to ensure that non-engineering students can benefit from them.
A close look at the literature shows the variety of techniques introduced for the fabrication of microfluidic structures without the use of conventional microfabrication techniques. This includes templating of micro-scale structures such as optical fibres,25 liquid metal,26,27 metal wires,28,29 and nylon threads30 in polymers such as polydimethylsiloxane (PDMS), and the subsequent removal of such structures either manually, chemically, or thermally following the curing of the polymer. Inspired by these techniques, we tried other materials that were more accessible (especially in low socio-economic regions), relatively cheaper, safer, reconfigurable to enable both simple and complex microfluidic structures, and usable by students and researchers from various backgrounds.
In this manuscript, we developed a novel fabrication method to produce microfluidic structures in the classroom environment. This method involves only commercially available consumables, such as pasta and petroleum jelly. We were not limited to straight channels and were able to create more complex structures, with the addition of a few easy steps to our basic method. For instance, by utilising a rotary grinding tool, we were able to subtract sections of pasta to create a contraction in the straight channel. We also soaked pasta to deform it into curved structures such as serpentines as well as combined structures such as mixers or droplet generators. To further highlight the versatility of our method, we created high aspect ratio and twisted serpentine structures. We also produced structures with varying heights and a 3D spiral channel completely embedded in PDMS. We demonstrated the biocompatibility of our pasta templated devices by showing that endothelial cells can be grown under shear stress condition inside the microfluidic channel. Therefore, our “do-it-in-classroom” fabrication approach facilitates the development of interactive microfluidics courses and is suitable for students with non-engineering backgrounds.
MATERIAL AND METHODS
Fabrication of microfluidic structures via pasta templating
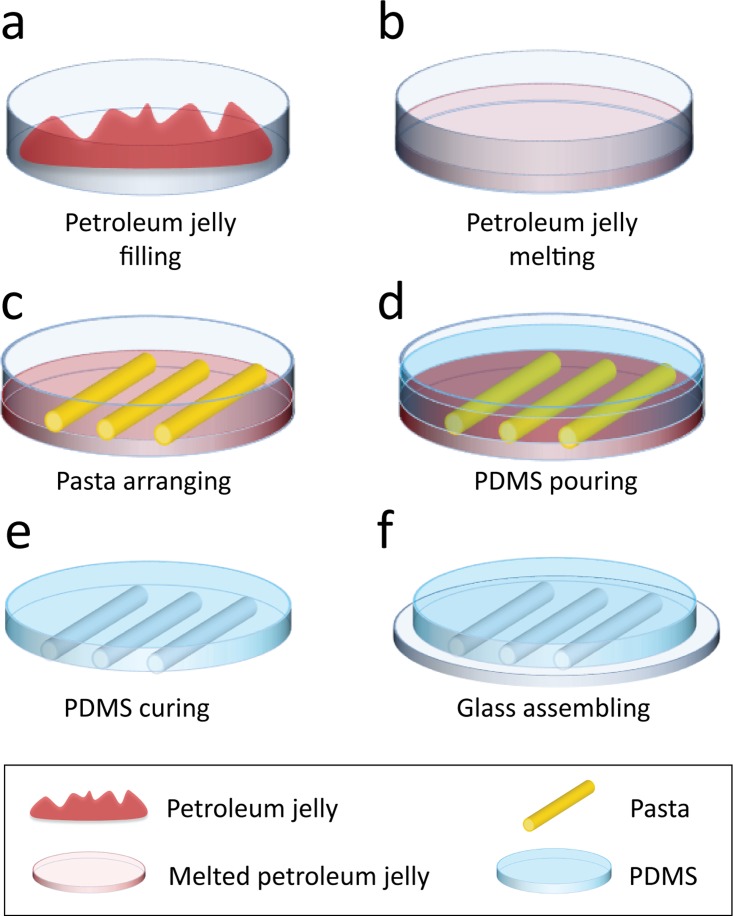
The process of fabrication is presented in Fig. 1 and Fig. S1 (supplementary material). A standard 85 mm diameter plastic Petri dish was filled with ∼18 ml of white petroleum jelly (Vaseline®) at room temperature [Fig. 1(a)]. The Petri dish was placed in a conventional toaster oven (Home & Co, Toaster Oven MG18CHV) at 70 °C for 15 min until the Petroleum jelly was melted and transparent [Fig. 1(b)]. The Petri dish was removed from the oven and pasta structures (San Remo) were manually patterned on the petroleum jelly surface as it cooled down (∼1.5 min after removal from oven) [Fig. 1(c)]. The petroleum jelly was left to cool down and transits into its gel phase (∼2 min). PDMS base and curing agent (Sylgard® 184 Silicone Elastomer Kit) were mixed at a standard 10:1 w/w ratio and poured onto the Petri dish [Fig. 1(d)]. The Petri dish was left on a level surface for the PDMS to cure at room temperature for 48 h.
FIG. 1.
Schematics of the replica moulding of pasta structures. (a) Petri dish is filled with petroleum jelly (Vaseline®). Petroleum jelly is shown in red for contrast. (b) Petroleum jelly is melted in a conventional oven at 70 °C for 15 min until transparent. (c) Pasta is patterned in desired configurations and placed onto the petroleum jelly before it transits into its gel phase. (d) PDMS is prepared at a 10:1 w/w ratio and poured onto the Petri dish. (e) PDMS is cured at room temperature (∼25 °C) for 48 h. After fully cured, PDMS is peeled off the Petri dish. (f) PDMS block with embedded microfluidic channels is assembled onto a glass slide and clamped to avoid leakage.
Our experiments indicated that uncured PDMS and petroleum jelly were immiscible. After curing, the PDMS block was peeled off the Petri dish [Fig. 1(e)]. The pasta structures generally remained in the petroleum jelly and could be easily removed to be reused. The PDMS blocks with embedded microfluidic structures were then cut into individual blocks with a scalpel to be assembled onto a 76 × 26 × 1 (L × W×H) mm glass slide (Menzel-Glaser) [Fig. 1(f)]. The PDMS blocks were punched with a 1.0 mm biopsy punch (Harris Uni-Core) at the inlet and outlet ports of the channel. A customised mechanical clamp made of 3 mm thick polymethyl methacrylate (PMMA) was used to seal the PDMS block against the glass slide, avoiding leakage at high flow rates. Eight 3 mm screws were used to tighten the clamp.
Experimental setup
Tygon® tubes (ID 0.02 and OD 0.06 in., 06419-01, Cole-Palmer®), assembled with aluminium tips cut from syringe needles (TERUMO, 23G), were used for interfacing the microfluidic structures with the syringe pump (Harvard Apparatus PHD 2000). For visualisation, water was stained with food dye (Queen) at a volumetric ratio of 5:1 (water:dye) and infused through the structures at varying flow rates of 5 to 2000 μl/min. For particle migration experiments, we used 8 and 15 μm polystyrene particles (PPs-8.0, PPs-15.0, Kisker Biotech GmbH & Co. KG) for brightfield imaging and yellow 7.3 μm fluorescent particles (PFP-6052, Kisker Biotech GmbH & Co. KG) for fluorescent imaging.
The channels were observed using a USB (Universal Serial Bus) microscope (Celestron COSMOS 5MP LCD Digital Microscope). An inverted microscope (Nikon Eclipse, TE 2000) equipped with a cooled CDD camera (QuantEM:512SC, Photometrics) and a 10× objective (CFI Plan Apo Lambda 10×) was used for high-resolution imaging. The blue fluorescence filter (Nikon B-2A, 450–490 nm) was used for observing the yellow fluorescent particles. For studying surface details of PDMS, we sputtered the PDMS channels with a thin 20 nm layer of gold and observed the surface under a scanning electron microscope (FEI Quanta) coupled with a back-scattered secondary electron Everhart-Thornley detector operated under high voltage and high vacuum. Temperature measurements along the surface of the petroleum jelly were performed using an infrared camera (FLIR systems, Thermo Vision A320, Sweden) interfaced with the ThermaCAM researcher software.
Cell culture, overnight flow experiment, and immunocytochemistry
For cell culture, the microfluidic channel was mounted on a glass cover slip using a mechanical clamp. The channel was washed with 70% ethanol followed by sterile water by applying a flow rate of 1 ml/min through the channel for 30 min using a syringe pump (Harvard Apparatus PHD 2000). The channel was then coated with human extracellular matrix components (MaxGelTM ECM, Sigma-Aldrich) according to supplier's instruction. Human Umbilical Vein Endothelial Cells (HUVECs) were purchased from Lonza and cultured at a density of 2.5 × 106 cell/ml of media inside the channel using EGMTM–2 growth media supplemented with SingleQuotsTM kit (Lonza) overnight to form a complete monolayer. Next, the cell growth medium was recirculated through the channel at a flow rate of 1.25 ml/min for 24 h using a peristaltic pump (OINA QP6 LAB High Accuracy) to induce a shear stress of 1 dyn/cm2 on the cultured cells. To examine the remodelling of cytoskeleton and formation of actin stress fibre in the cultured cells, the cells were fixed with 4% paraformaldehyde for 1 h and permeabilised with 0.2% Triton X-100 in PBS (phosphate-buffered saline) for 30 s. Nonspecific binding was blocked with 5% bovine serum albumin in PBS and probed with phalloidin that was directly conjugated to Alexa Fluor 568 (Sigma-Aldrich) and DAPI 4′,6-diamidino-2-phenylindole (Sigma-Aldrich). Imaging was performed on a Nikon A1 laser scanning confocal microscope.
Computational fluid dynamics (CFD)
CFD was used to simulate the characteristics of each structure such as flow velocity and particle tracking. Accurate 3D models of fabricated structures were constructed in Fusion360 CAD software by recreation and extrusion of channel cross-sections taken by SEM imaging. In general, the geometry of structures remained unchanged after clamping. In the case of major deformations after clamping (e.g., occurring for high aspect ratio serpentine channels), macro images of deformed cross-sections were taken using a digital single-lens reflex camera (Canon 6D equipped with a Canon EF 100 mm f/2.8 macro ultrasonic motor lens) to be recreated and extruded in Fusion360. The 3D models were then exported to Gambit-2.3 (ANSYS Fluent) for mesh generation. Fluent-6.3 (ANSYS Fluent) was used for simulating and post-processing of velocity contours and particle tracking through the microfluidic structures. Second-order upwind model accompanied with SIMPLE-C algorithm was used for coupling of velocity and pressure. Boundary conditions included desired flow rates at the inlet, zero pressure at the outlet, and no slip at the walls and surface of the glass slide.
RESULTS AND DISCUSSIONS
Straight channels with controllable semi-circular cross-sections
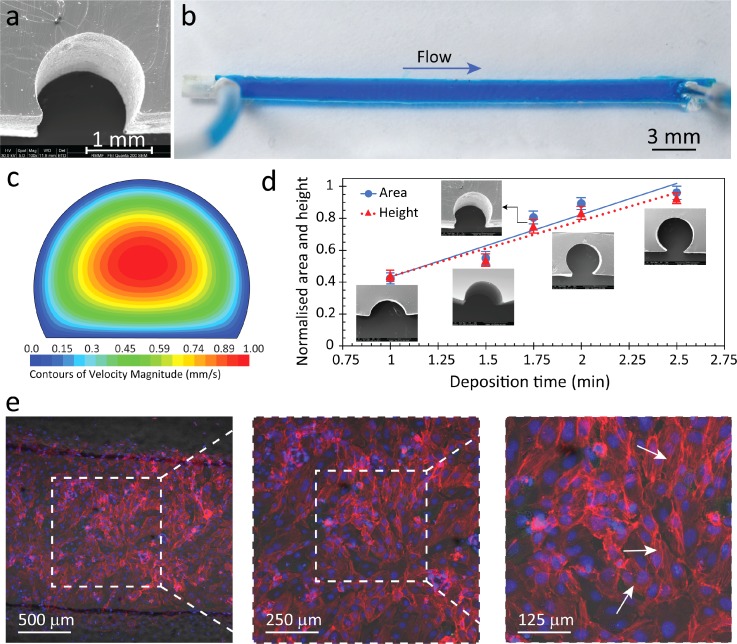
In its simplest form, our method was capable of fabricating straight channels with semi-circular cross-sections [Fig. 2(a)] by arranging uncooked 1.5 mm diameter cylindrical pasta on top of petroleum jelly. We utilised pasta as our channel component because it retains its shape when dry, and it can also be deformed when wet. Our previous attempts with thin copper and aluminium wires were undesirable as the wires would wrinkle and not lay flat. We had also tried to place pasta directly onto the Petri dish and filled it with PDMS. However, this resulted in a thin skin being formed at the bottom of the channel, which obstructs viewing of channel contents (see supplementary material Fig. S2). We chose petroleum jelly as the base material because it transits from liquid to gel phase in a matter of minutes, it is safe and easy to handle, and biocompatible. Other substances such as wax, honey, and chocolate were also used as the base material; however, the resultant PDMS would not adhere to glass, had wrinkles at the edges of the channel (see supplementary material Fig. S3), or was too dirty to operate. A flat bottom surface in the channel eases the observation of samples flowing through the channel when using an inverted microscope and the round top surface mimics the structure of blood vessels. Interestingly, scanning electron microscopy (SEM) imaging also showed a rough interior surface [see supplementary material Figs. S4(a) and S4(b)], which also resembles the rough surface of blood vessels due to the presence of endothelial cells.31–33 Although petroleum jelly slightly changed the surface roughness of PDMS [see supplementary material Fig. S4(c)] and reduced its adhesiveness, we were able to overcome these limitations by mechanically clamping PDMS to glass, eliminating any leakage [Fig. 2(b)].
FIG. 2.
Straight channel with a semi-circular cross-section fabricated via pasta templating. (a) High-resolution microscopy of the channel cross-section obtained via SEM. (b) Top view of the PDMS channel filled with blue stained water for improved visualisation, indicating no leakage. (c) Velocity contours of fluid at the cross-section of the channel obtained by numerical simulations at a flow rate of 50 μl/min. (d) Cross-sectional area and height against deposition time. Solid line indicates area, while dotted line indicates height. Error bars correspond to average ± standard deviation obtained from four sets of independent fabricated structures. (e) HUVECs were grown inside the channel under a flow induced shear stress of 1 dyn/cm2 for 24 h. F-actin was stained with Alexa Fluor 568 (red), while nuclei were stained with DAPI (blue). White arrows indicate the formation of actin stress fibres in endothelial cells when grown under shear stress.
To verify the leakage-free performance of the channel, we infused blue stained water into the channel at various flow rates of 5 to 2000 μl/min for at least 30 min. Velocity contours obtained via CFD simulations at a flow rate of 50 μl/min showed a maximum velocity of 1 mm/s at the centre of the channel and zero velocity at the walls, as expected [Fig. 2(c)]. The Reynold's number based on the hydraulic diameter of the channel was calculated as (in which ρ and μ are the density and viscosity of the liquid, U is the average velocity, and Dh is the hydraulic diameter of the channel cross-section), which indicates laminar characteristics of flow. The leakage free performance of the channel was also verified by applying a solution of water and glycerol that was 20 times more viscous than water at flow rates ranging from 1 to 200 μl/min.
A unique feature of our fabrication method entails the utilisation of a petroleum jelly (Vaseline®) substrate rather than a conventional solid substrate. This petroleum jelly is a temperature-sensitive colloidal system that undergoes a liquid-to-gel phase transition when cooling. Our experiments indicated that this phase change occurs from 59 to 48 °C within ∼0.75–2 min when cooling by natural convection mechanism at room temperature and was associated with colour change of the petroleum jelly (see supplementary material Fig. S5 + Video S1). The liquid-to-gel phase transition changed the surface properties of the substrate and subsequently changed the cross-sectional area of the pasta submerged in the petroleum jelly. This enabled us to modulate the cross-sectional area and height of the channels by simply changing the deposition time (see supplementary materials Figs. S6 and S7) of the pasta onto the petroleum jelly [Fig. 2(d)]. As shown, the cross-sectional area of the channel can range from 42% to 96% of the total cross-sectional area of the pasta, and from 44% to 93% of the total cross-sectional height. This means that we can achieve cross-sections that are only a segmental fraction of a circle, to an almost complete circle, as the pasta is submerged in the petroleum jelly at different levels. However, even after transitioning into its gel phase, the petroleum jelly surface was soft enough to allow the pasta to be submerged ∼100 μm in the jelly, limiting the maximum cross-sectional area to ∼96%. A 100% cross-sectional area could be achieved in the absence of the petroleum jelly layer, but this led to the formation of a thin skin of PDMS underneath the channel, which was undesirable for inverted microscopy imaging. This experiment clearly shows increased versatility compared to conventional rectangular shaped channels made from lithography.
To confirm the biocompatibility of our channels for conducting cellular experiments, endothelial cells (HUVECs) were cultured in a channel, as detailed in Materials and Methods. The cell growth medium was recirculated through the channel at a flow rate of 1.25 ml/min (corresponding to a shear stress of 1 dyn/cm2 induced on the cultured cells) for 24 h. The response of HUVECs to shear stress was analysed in terms of the remodelling of cytoskeleton and formation of actin stress fibres, as detailed in Materials and Methods [Fig. 2(e)]. The results indicated the formation of thin stress fibres (indicated by white arrows) distributed across the cell as a result of shear stress.34–36 In comparison, control experiments in no-flow conditions indicated the formation of dense actin bands only at the periphery of cells (see supplementary material Fig. S8). The results suggest the suitability of our channels for conducting cell-based assays.37–40
Straight channels with local contractions
We also demonstrated the ability to create arbitrary geometries using subtractive methods. As a proof-of-concept, we made a straight channel with a semi-circular contraction using a rotary grinding tool (Dremel Micro) equipped with a 3 mm ball shape rotary burr at 15 000 rpm. Flat pasta with an elliptical cross-section was used as its larger width allows for more control over the size of the contraction as well as better visualisation. Although similar contracted structures can be obtained with the circular pasta used previously for the straight channel, if desired. Microfluidic channels with local contractions are commonly used for the hydrodynamic manipulation (focusing, size sorting) of microparticles.41 Also, such contractions can mimic an anomalous narrowing of blood vessels known as a stenosis, for studying shear induced platelet aggregation.42
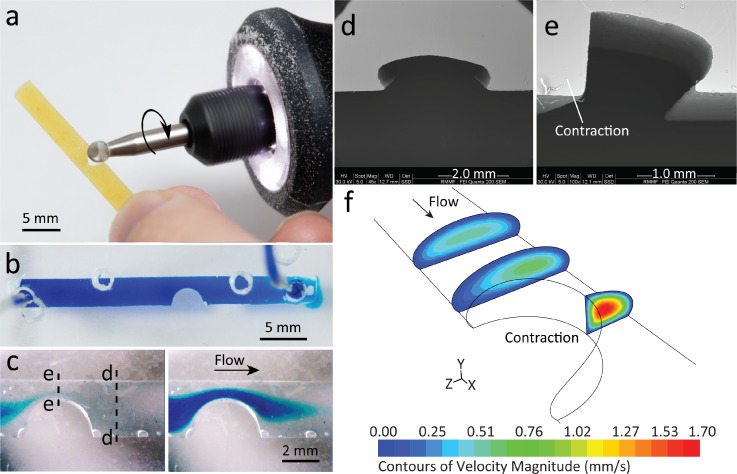
In order to create narrowed channels, we used a rotary grinding tool to subtract sections of the pasta manually within 2 min [Fig. 3(a)]. This process resulted in a semicircular contraction of 3.65 mm in length with a contraction ratio of , where W is the width. We were able to control the contraction ratio from 20% to 80% by adjusting the amount of subtracted pasta, meaning it was fully customisable. With the gentle removal of subtracted pasta from the PDMS structure, it can be reused for repeatable fabrication. Experiments showed that contraction ratios beyond 80% could potentially break the structure as the pasta becomes more fragile. However, one might use a customised clamping device to carefully hold the pasta during the grinding process and increase this threshold.
FIG. 3.
Channels with a semi-elliptical cross-section and a localised contraction fabricated via pasta templating. (a) Grinding tool subtracts a section of the pasta to create a localised contraction. (b) Top view of PDMS channel filled with blue stained water, emphasising the contracted shape with no leakage. (c) Snapshots of the filling process with blue stained water in a pre-filled water channel, showing flow acceleration at the narrowed passage. Dashed lines indicate point of dissection of the channel, to show the (d) semi-elliptical cross-section outside the contraction and (e) quarter-elliptical cross-section at the apex of the contraction. (f) Velocity contours of flow at various cross-sections of the channel, obtained at a flow rate of 50 μl/min.
The resultant contracted PDMS structure was adhered to glass via mechanical clamping between two sheets of PMMA with drilled access ports for the inlet/outlet and filled with blue stained water, which showed minimal leakage [Fig. 3(b)]. Additional holes were visible as the same mechanical clamp was used for a variety of structures. A video taken via a USB microscope (see supplementary material Video S2) captured the injection of blue stained water passing through the narrowed passage at a flow rate of 50 μl/min [Fig. 3(c)]. We observed that the stained water had an affinity towards the narrowed walls before filling out the entire channel due to the increased velocity at the site of the contraction.
SEM images show how the cross-section changes from semi-elliptical to quarter-elliptical when along the contracted structure [(Figs. 3(d) and 3(e)]. This explains the increased velocity as the cross-sectional area decreases, as shown in the numerical simulations [Fig. 3(f)]. The simplicity and time efficiency of this method facilitate the fabrication of more complex structures with multiple contractions along the same channel.
Curved and multi-inlet microfluidic channels
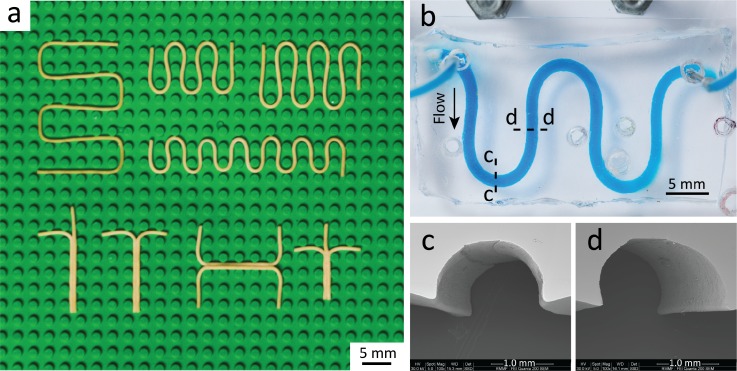
We further explored the versatility of our method for producing curved and multi-inlet structures. Pasta was soaked in lukewarm water (40 °C) for ∼45 min until soft and malleable (indicated by change in colour from yellow to pale yellow and expanded diameter) to create customised curved structures. We referred to this procedure as the soaking-drying process. LEGO® baseplates were utilised to shape the pasta into the desired arrangement as it was accessible, affordable, and produced consistent structures. For the case of serpentine structures, single pasta strands were placed around the pillars of the LEGO® baseplate in a zig-zag pattern. Multiple pasta strands were combined to create multi-inlet channels that could be operated as mixers, droplet generators, or H-shaped filters (Fig. 4). This process allowed parallel fabrication of multiple pasta structures on a single LEGO® baseplate. We could also deform the pasta by sandwiching it between two LEGO® baseplates to create channels with a more rectangular cross-section. As shown [Fig. 4(a)], pasta was left to air dry for 3 h in its desired formation until rigid (indicated by the colour reversion to a darker yellow and diameter shrinkage) before it was removed from the LEGO® baseplate and implemented in the previously established pasta templating method. Although the drying process can be achieved instantly via hot air, experiments show that this leads to thermal bending.
FIG. 4.
Customised curved structures fabricated via pasta templating. (a) Soaked pasta placed in customised arrangements on a LEGO® baseplate. Top row shows serpentine structures of varying sizes. Bottom row shows multi-inlet structures using multiple pasta strands in y-, T-, H-, and t-shaped channels, respectively. (b) Top view of serpentine channel in PDMS filled with blue stained water. Dashed lines show dissection point for the following cross-sections. SEM shows consistent semi-circular cross-sections both at the (c) curved and (d) straight sections of the serpentine channel.
A serpentine channel patterned in PDMS was adhered to glass via mechanical clamping and filled with blue stained water at 50 μl/min, showing no leakage [Fig. 4(b)]. Serpentine channels are common structures in microfluidics due to its increased length, which is advantageous in enhancing passive mixing or increasing reaction time.43 The Dean number was calculated as (where W is the width of the channel and R is the radius of one curvature of the serpentine), which shows that Dean vortices are not dominant.44 By using the LEGO® baseplate, we were able to produce serpentines, which were quite uniform and had curves that were equidistant apart. The cross-section was also shown to be consistent throughout the channel, even after the soaking-drying process [Figs. 4(c) and 4(d)]. Although we have used commercially available LEGO®, any arbitrary building blocks containing arrays of pillars can be used for making customised or more complex curved structures.
Multi-inlet microfluidic structures enabling passive mixers and droplet generators
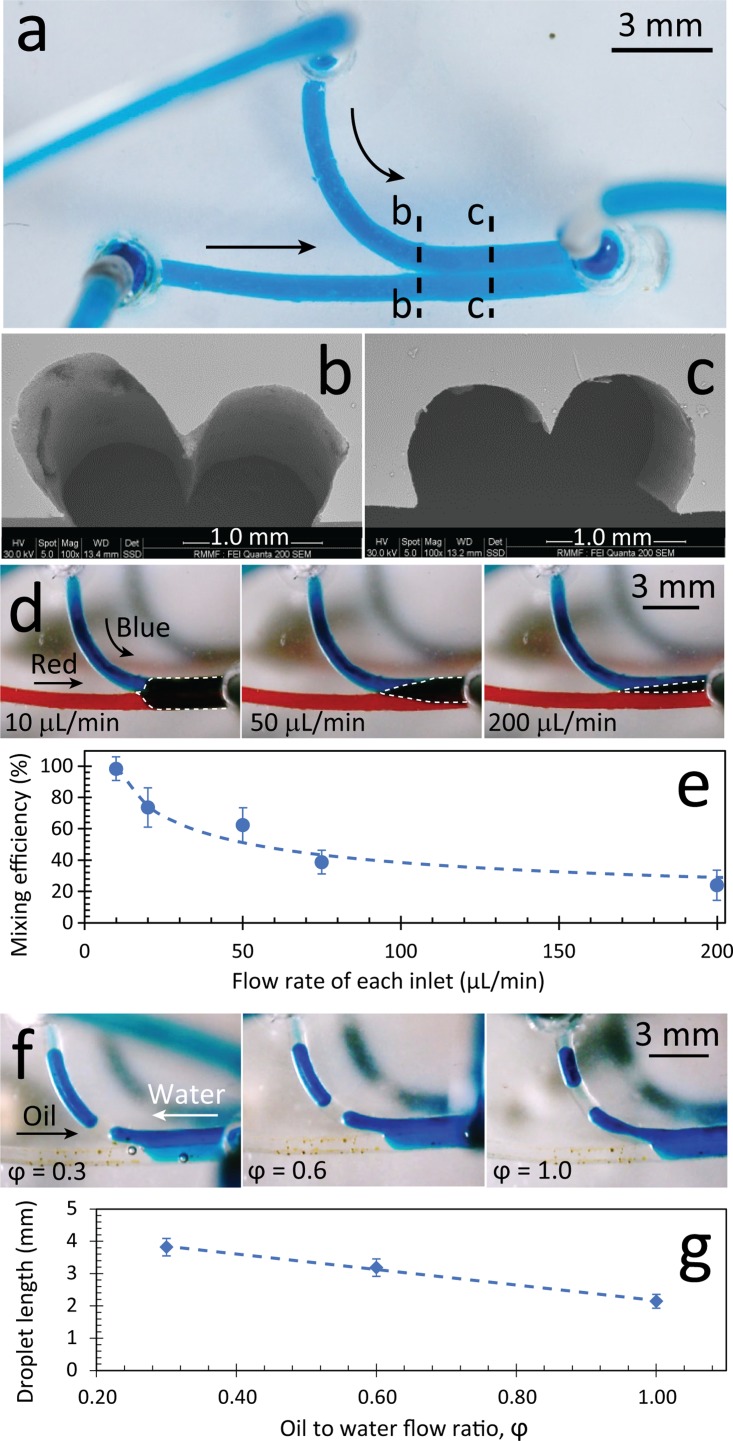
The y-channel created from the soaking-drying process was made with two inlets and one outlet to be used as both a mixer and a droplet generator [Fig. 5(a)]. The PDMS block was adhered to glass via mechanical clamping, as explained previously. SEM images showed the formation of a double arc cross-section at the downstream of the y-channel with a tapered wedge structure formed at the interface of the neighbouring channels [Figs. 5(b) and 5(c)]. Further SEM analysis indicated some surface irregularities along this wedge structure (see supplementary material Fig. S9), which may have been caused when the two neighbouring pasta strands were combined.
FIG. 5.
Double-inlet y-channel fabricated with semi-circular cross-sections via pasta templating. (a) Top view of PDMS structure filled with blue stained water for improved visualisation. SEM shows cross-sections at both the (b) junction of the two channels and (c) at the downstream. (d) Operation of the y-channel as a passive mixer by applying red and blue stained water at various inlet flow rates. The purple delta mixing region is shown by dashed lines. (e) Mixing efficiency vs. inlet flow rates. (f) Operation of the y-channel as a droplet generator by applying immiscible water and oil flows shown at various flow ratios of oil to water. (g) Droplet length vs. oil to water flow ratio. Results are obtained from five independent experiments, and shown as average ± standard error.
A pair of miscible liquids (i.e., red and blue stained water) was injected into the two inlets to investigate the passive mixing. We found immediate mixing between the neighbouring liquids, as evidenced by the formation of the purple delta region at the downstream of the channel [Fig. 5(d)]. Increasing the flow rate from 10 to 200 μl/min at each inlet significantly decreased the purple delta region and consequently reduced the mixing efficiency of neighbouring liquids [Fig. 5(e) + supplementary material Fig. S10]. Therefore, the y-channel operated as an effective passive mixer, in which the mixing efficiency could be controlled from 24% to 98%, dependent on the inlet flow rate. The Reynold's number was calculated as and the Péclet number was calculated as (where D is the mass diffusivity of food dye in water solutions) showing the dominance of diffusive mixing. We hypothesise that the strong passive mixing in our y-channel is due to the tapered wedge structure and conjugate surface irregularities (shown in Fig. S9). If undesired, surface irregularities can be removed by implementing some extra surface treatment steps, such as polishing the interface of the two pastas with fine polishing rotary tools or coating the undesired cavities along the interface with a thin layer of petroleum jelly.
Next, we applied immiscible liquids (i.e., water and olive oil) to study the utility of the same y-channel for the continuous generation of droplets (see supplementary material Video S3). Videos were taken with a USB microscope at 30 fps. Experiments showed that applying the water and oil under the same conditions shown in the passive mixer does not produce enough shear stress to pinch off the water into small droplets. We overcame this limitation by changing the positions of the inlets and outlet. As such, the two liquids would compete to enter the mixing channel, and sufficient shear stress was produced to pinch off the water into small droplets [Fig. 5(f)]. We continued this experiment for 30 min to ensure that there were no significant differences between the droplet sizes and droplet generation rates. The length of water droplets was inversely proportional to the flow ratio of oil to water, defined as .45 Hence, by increasing from 0.3 to 1.0, we were able to reduce the length of the droplets from 4 to 2 mm [Fig. 5(g)]. Consequently, this reduced the generation rate of droplets from 1.5 to 0.2 droplets per second. The generation rate of droplets could be amplified by increasing the flow rates of oil and water at the same flow ratio.
Microfluidic channels with unconventional cross-sections
We also examined the versatility of our fabrication method for creating microfluidic structures with unconventional cross-sections. As a proof-of-concept, we fabricated high aspect ratio and twisted serpentine channels. We produced a serpentine channel with a high aspect ratio by adopting flat pasta in the soaking-drying process and placing the soft pasta on its thinner side in the LEGO® baseplate [Fig. 6(a)]. SEM imaging indicated an oblong cross-section in the channel [Fig. 6(b)]. However, the increased complexity created bowing at the bottom edge of the microfluidic channel. This bowing may be due to the formation of a convex meniscus on the surface of the petroleum jelly layer interfaced with the high aspect ratio pasta structures and PDMS. The channel deformation caused by clamping led to an even surface at the interface of the serpentine channel and the glass slide, which was desired for achieving leakage-free flow [Fig. 6(c)]. Numerical simulations using this modified clamped cross-section predicted a maximum flow velocity of 0.7 mm/s towards the wider region of the cross-section [Fig. 6(d)]. No leakage was observed at flow rates ranging from 5 to 2000 μl/min, indicating the effectiveness of our clamping technique [Fig. 6(e)]. This confirms the utility of our pasta templating method for producing high aspect ratio structures, which is challenging using conventional lithographic processes. Such high aspect ratio structures can be particularly interesting for sorting of particles based on dimensions or density by applying magnetic or electrical forces.46
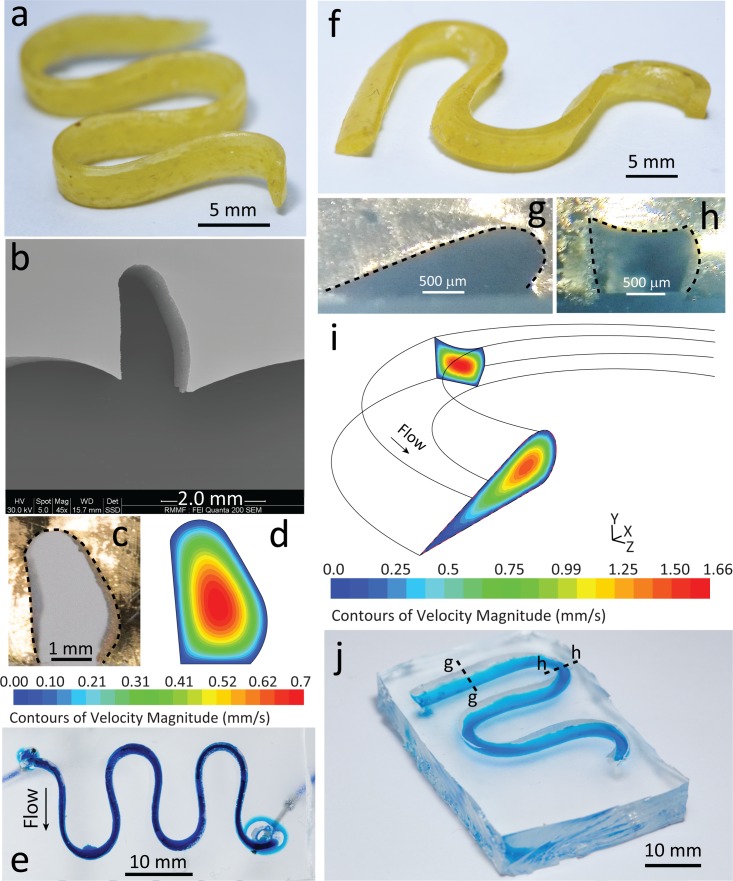
FIG. 6.
3D microfluidic structures with unconventional cross-sections fabricated from flat pasta via pasta templating. (a) Pasta arranged in a serpentine formation, to produce a channel with high aspect ratio. (b) SEM shows semi-oblong cross-section and bowing at the bottom of the channel. (c) Cross-section of channel after mechanical clamping. (d) Flow velocity contours based on the clamped cross-section obtained at 50 μl/min. (e) Bottom view of high aspect ratio channel filled with blue stained water for better visualisation. (f) Pasta arranged in a serpentine formation deformed at a 45° angle, to produce a twisted serpentine structure. Cross-sections both at the (g) curved and (h) straight sections of the structure after clamping. (i) Velocity contours at the prescribed cross-sections obtained at 50 μl/min. (j) Isometric view of twisted serpentine structure filled with blue stained water, to emphasise its 3D nature. Dashed lines indicate point of dissection for the previous cross-sections.
The twisted serpentine channel was made by adopting the same procedure explained above and then compressing the pasta structure using a second LEGO® baseplate on top, until it was in contact with the bottom baseplate [Fig. 6(f)]. The twisted channel exhibited variable cross-sections at different points in the channel. The cross-section drastically transformed from an aerofoil shape at the straight section of the serpentine [Fig. 6(g)] to a rectangular shape at the sharp turns of the serpentine [Fig. 6(h)]. This resulted in significantly different flow profiles at different locations of the channel as evidenced by numerical simulations [Fig. 6(i)], which were based on the clamped cross-sections (see supplementary material Fig. S11). The PDMS channel was filled with blue stained water to visualise its 3D twisted effect as shown by the gradient of blue at the straight sections of the channel and the concentrated dark blue at the curves [Fig. 6(j)]. This irregular structure could be used to observe the dynamics of particles under varying channel cross-sections.47
More complex 3D structures embedded in PDMS
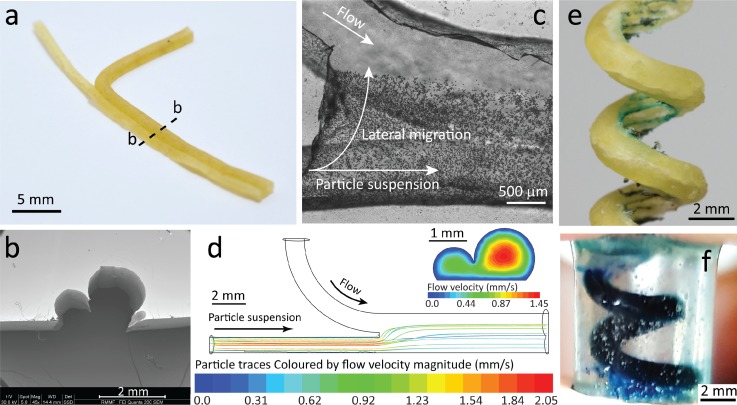
Next, we explored the utility of the pasta templating method for fabrication of more complex microfluidic structures. First, we fabricated a y-channel with varying channel diameters by combining a thin pasta strand (Ø 0.8 mm) with a thick pasta strand (Ø 1.5 mm) [Fig. 7(a)], which can be clearly seen in SEM images [Fig. 7(b)]. This enabled us to study the lateral migration of particles across the channel. By injecting 15 μm polystyrene particles into the smaller channel, we observed the lateral particle migration toward the larger channel immediately after the junction [Fig. 7(c)]. The vast change in channel diameters induced sufficient hydrodynamic lift force48 drawing the particles towards the larger channel. This was also verified through numerical simulations [Fig. 7(d)]. The rendered view of the y-channel structure used in our simulations is presented in Fig. S12 (supplementary material). This phenomenon was further demonstrated with smaller 8 μm particles and 7.3 μm fluorescent particles (see supplementary material Video S4 + Video S5). Increasing the flow rate of water through the larger channel leads to complete migration of particles at the end of the channel (see supplementary material Video S4). Such structures can be useful for studying the lateral migration of bio-particles under hydrodynamic forces.49
FIG. 7.
More complex 3D structures fabricated via pasta templating. (a) Two pasta strands of varying diameter (Ø 0.8 mm and Ø 1.5 mm) combined to create a y-channel. Dashed line indicates point of dissection for the following cross-section. (b) SEM shows the difference in height in the cross-section at the downstream of the y-channel. (c) High-resolution stitched image, via inverted microscopy, showing 15 μm particles injected in the small channel, migrating laterally towards the large channel. (d) Particle tracking simulations show the migration of the particles at the junction of the y-channel. Particle traces are coloured according to the velocity magnitude. Inset shows flow velocity contours at the downstream with obtained at 50 μl/min at each inlet. (e) Pasta formed into a spiral structure via a 3 mm screw. Threading inside the spiral was made visible with blue stained water. (f) Side view of spiral channel embedded in a cylindrical block of PDMS, filled with blue stained water for visualisation.
Finally, we created a 3D spiral channel completely encapsulated in PDMS by coiling a soaked pasta strand around a 3 mm screw [Fig. 7(e)]. The pasta was then left to dry and was removed from the screw. The resultant spiral structure was placed in a 1.5 ml Eppendorf tube. PDMS was poured into the tube to submerge the pasta and after 48 h, produced a cylindrical block of PDMS with an embedded pasta spiral. To remove the pasta, we soaked the block of PDMS in 1M sodium hydroxide (NaOH). NaOH was able to gradually deteriorate the pasta to develop an empty spiral channel in PDMS. Manual compression every hour (up to a total of 6 h) was necessary to remove the disintegrated parts of the pasta and ensure that the entire embedded pasta would be exposed to the NaOH solution. After all the pasta fragments were removed, the spiral channel was flushed with water at 120 μl/min for 10 min to eliminate any traces of NaOH. This empty spiral channel was then filled with blue stained water for visualisation purposes, indicating no leakage [Fig. 7(f)]. Spiral structures can be used to enhance passive mixing by inducing Dean vortices44,50 as well as making microfluidic based electronic devices.26,51 This sacrificial moulding technique26,51 facilitates the fabrication of multilayer PDMS channels useful for organ-on-chip devices.52–54
CONCLUSIONS
We have established a novel process for the fabrication of micro- to milli-scale microfluidic structures. This so-called “do-it-in-classroom” fabrication method involved only affordable and accessible consumables and equipment, as shown by the use of pasta, petroleum jelly, and a conventional oven. We were able to fabricate conventional structures used in microfluidics (straight channels, serpentines, mixers, and droplet generators) as well as unconventional structures (high aspect ratio serpentines, twisted serpentines, y-channels with varied cross-sectional heights, and 3D spiral channels).
With our method, the height of the channel can be conveniently controlled by varying the deposition time, enabling controllable cross-sectional profiles from semi-circular to almost entirely circular. Also, the user is not limited to the geometry and structure of the pasta, as it can be changed using subtractive methods as well as the soaking-drying process for customised configurations, as demonstrated. Fabrication of multilayer structures useful for producing organ-on-chip devices52 is also possible by removing the pasta with NaOH. Furthermore, pasta with smaller diameters can also be used to create smaller channels and the surface roughness of the pasta can be manipulated while pasta is soft.
Our method does not offer the precision, resolution, and surface quality (smoothness) of commonly used microfabrication technologies.55 Despite these drawbacks, it enables the fabrication process to be conducted entirely in the classroom.56 This method also combines the design and fabrication steps and therefore is suitable for non-engineers who do not necessarily have sufficient CAD knowledge. With our method, we can bridge the gap between the classroom and the laboratory, facilitating project-based microfluidic courses that promote creative and critical thinking skills that are highly desired in the current workforce.
Together with other manually operated self-sufficient microfluidic systems,57–60 the pasta templating method can facilitate the exposure of biologists and practitioners to the emerging field of microfluidics. Furthermore, our method facilitates the expansion of the field of microfluidics in institutions and countries that do not have access to microfabrication facilities but wish to be involved in the emerging market of microfluidics.
SUPPLEMENTARY MATERIAL
See supplementary material for detailed information of methods, additional experimental data, and videos showing the liquid-to-gel phase transition of petroleum jelly as well as operation of fabricated chips for the localised contracted channel, droplet generation, and particle lateral migration.
ACKNOWLEDGMENTS
Authors wish to acknowledge RMIT Microscopy and Microanalysis Facility (RMMF) for scanning electron microscopy (SEM) imaging of fabricated structures. N.N. acknowledges RMIT University School of Engineering for Ph.D. Scholarship. E.P. acknowledges National Health and Medical Research Council for funding “The Australian Centre for Electromagnetic Bioeffects Research” (NHMRC CRE APP1135076). S.B. acknowledges financial support from the Australian Research Council (DE170100239). K.K. acknowledges financial support from the Australian Research Council (DP180102049).
The authors declare no competing financial interest.
References
- 1. Chin C. D., Laksanasopin T., Cheung Y. K., Steinmiller D., Linder V., Parsa H., Wang J., Moore H., Rouse R., Umviligihozo G., Karita E., Mwambarangwe L., Braunstein S. L., Van De Wijgert J., Sahabo R., Justman J. E., El-Sadr W., and Sia S. K., Nat. Med. 17(8), 1015–1019 (2011). 10.1038/nm.2408 [DOI] [PubMed] [Google Scholar]
- 2. Lai S., Wang S., Luo J., Lee L. J., Yang S. T., and Madou M. J., Anal. Chem. 76(7), 1832–1837 (2004). 10.1021/ac0348322 [DOI] [PubMed] [Google Scholar]
- 3. Baratchi S., Khoshmanesh K., Sacristán C., Depoil D., Wlodkowic D., McIntyre P., and Mitchell A., Biotechnol. Adv. 32(2), 333–346 (2014). 10.1016/j.biotechadv.2013.11.008 [DOI] [PubMed] [Google Scholar]
- 4. LaVan D. A., McGuire T., and Langer R., Nat. Biotechnol. 21(10), 1184–1191 (2003). 10.1038/nbt876 [DOI] [PubMed] [Google Scholar]
- 5. Moghadas H., Saidi M. S., Kashaninejad N., and Nguyen N.-T., Biomicrofluidics 12(2), 024117 (2018). 10.1063/1.5021002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prescient & Strategic (P&S) Market Research, Microfluidic Devices Market Overview, https://www.psmarketresearch.com/market-analysis/microfluidic-devices-market [Google Scholar]
- 7. Weibel D. B., DiLuzio W. R., and Whitesides G. M., Nat. Rev. Microbiol. 5, 209 (2007). 10.1038/nrmicro1616 [DOI] [PubMed] [Google Scholar]
- 8. Razzacki S. Z., Thwar P. K., Yang M., Ugaz V. M., and Burns M. A., Adv. Drug Delivery Rev. 56(2), 185–198 (2004). 10.1016/j.addr.2003.08.012 [DOI] [PubMed] [Google Scholar]
- 9. Blanco F. J., Agirregabiria M., Garcia J., Berganzo J., Tijero M., Arroyo M. T., Ruano J. M., Aramburu I., and Kepa M., J. Micromech. Microeng. 14(7), 1047 (2004). 10.1088/0960-1317/14/7/027 [DOI] [Google Scholar]
- 10. Becker H. and Gärtner C., Electrophoresis 21(1), 12–26 (2000). [DOI] [PubMed] [Google Scholar]
- 11. Morelli L., Serioli L., Centorbi F. A., Jendresen C. B., Matteucci M., Ilchenko O., Demarchi D., Nielsen A. T., Zor K., and Boisen A., Lab Chip 18(6), 869–877 (2018). 10.1039/C7LC01217A [DOI] [PubMed] [Google Scholar]
- 12. Lee U. N., Su X., Guckenberger D. J., Dostie A. M., Zhang T., Berthier E., and Theberge A. B., Lab Chip 18(3), 496–504 (2018). 10.1039/C7LC01052D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heckele M. and Schomburg W. K., J. Micromech. Microeng. 14(3), R1–R14 (2004). 10.1088/0960-1317/14/3/R01 [DOI] [Google Scholar]
- 14. Thompson B. L., Ouyang Y., Duarte G. R. M., Carrilho E., Krauss S. T., and Landers J. P., Nat. Protoc. 10(6), 875–886 (2015). 10.1038/nprot.2015.051 [DOI] [PubMed] [Google Scholar]
- 15. Su X., Zhang S., Ge S., Chen M., Zhang J., Zhang J., and Xia N., Biomicrofluidics 12(2), 024108 (2018). 10.1063/1.5019650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ho C. M. B., Ng S. H., Li K. H. H., and Yoon Y.-J., Lab Chip 15(18), 3627–3637 (2015). 10.1039/C5LC00685F [DOI] [PubMed] [Google Scholar]
- 17. Waheed S., Cabot J. M., Macdonald N. P., Lewis T., Guijt R. M., Paull B., and Breadmore M. C., Lab Chip 16(11), 1993–2013 (2016). 10.1039/C6LC00284F [DOI] [PubMed] [Google Scholar]
- 18. Park Y.-J., Yu T., Yim S.-J., You D., and Kim D.-P., Lab Chip 18, 1250–1258 (2018). 10.1039/C8LC00004B [DOI] [PubMed] [Google Scholar]
- 19. Lee Y.-S., Bhattacharjee N., and Folch A., Lab Chip 18, 1207–1214 (2018). 10.1039/C8LC00001H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kang K., Oh S., Yi H., Han S., and Hwang Y., Biomicrofluidics 12(1), 014105 (2018). 10.1063/1.5012548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Waldbaur A., Rapp H., Länge K., and Rapp B. E., Anal. Methods 3(12), 2681–2716 (2011). 10.1039/c1ay05253e [DOI] [Google Scholar]
- 22. Li F., Smejkal P., Macdonald N. P., Guijt R. M., and Breadmore M. C., Anal. Chem. 89(8), 4701–4707 (2017). 10.1021/acs.analchem.7b00409 [DOI] [PubMed] [Google Scholar]
- 23. Tovar M., Weber T., Hengoju S., Lovera A., Munser A.-S., Shvydkiv O., and Roth M., Biomicrofluidics 12(2), 024115 (2018). 10.1063/1.5013325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kadilak A. L., Rehaag J. C., Harrington C. A., and Shor L. M., Biomicrofluidics 11(5), 054109 (2017). 10.1063/1.5003477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mannino R. G., Myers D. R., Ahn B., Wang Y., Margo R., Gole H., Lin A. S., Guldberg R. E., Giddens D. P., Timmins L. H., and Lam W. A., Sci. Rep. 5, 12401 (2015). 10.1038/srep12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parekh D. P., Ladd C., Panich L., Moussa K., and Dickey M. D., Lab Chip 16(10), 1812–1820 (2016). 10.1039/C6LC00198J [DOI] [PubMed] [Google Scholar]
- 27. Yan S., Li Y., Zhao Q., Yuan D., Yun G., Zhang J., Wen W., Tang S.-Y., and Li W., Lab Chip 18(5), 785–792 (2018). 10.1039/C8LC00047F [DOI] [PubMed] [Google Scholar]
- 28. Song S.-H., Lee C.-K., Kim T.-J., Shin I.-c., Jun S.-C., and Jung H.-I., Microfluid. Nanofluid. 9(2), 533–540 (2010). 10.1007/s10404-010-0570-y [DOI] [Google Scholar]
- 29. Venzac B., Madoun R., Benarab T., Monnier S., Cayrac F., Myram S., Leconte L., Amblard F., Viovy J.-L., Descroix S., and Coscoy S., Biomicrofluidics 12(2), 024114 (2018). 10.1063/1.5025027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Camargo C. L., Shiroma L. S., Giordano G. F., Gobbi A. L., Vieira L. C. S., and Lima R. S., Anal. Chim. Acta 940, 73–83 (2016). 10.1016/j.aca.2016.08.052 [DOI] [PubMed] [Google Scholar]
- 31. Schmidt-Trucksäss A., Sandrock M., Cheng D. C., Müller H. M., Baumstark M. W., Rauramaa R., Berg A., and Huonker M., Atherosclerosis 166(1), 57–65 (2003). 10.1016/S0021-9150(02)00245-9 [DOI] [PubMed] [Google Scholar]
- 32. Niu L., Qian M., Yang W., Meng L., Xiao Y., Wong K. K. L., Abbott D., Liu X., and Zheng H., PLoS One 8(10), e76880 (2013). 10.1371/journal.pone.0076880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsvirkun D., Grichine A., Duperray A., Misbah C., and Bureau L., Sci. Rep. 7, 45036 (2017). 10.1038/srep45036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pellegrin S. and Mellor H., J. Cell Sci. 120(20), 3491 (2007). 10.1242/jcs.018473 [DOI] [PubMed] [Google Scholar]
- 35. Noria S., Xu F., McCue S., Jones M., Gotlieb A. I., and Langille B. L., Am. J. Pathol. 164(4), 1211–1223 (2004). 10.1016/S0002-9440(10)63209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu M.-C., Shih H.-C., Wu J.-G., Weng T.-W., Wu C.-Y., Lu J.-C., and Tung Y.-C., Lab Chip 13(9), 1743–1753 (2013). 10.1039/c3lc41414k [DOI] [PubMed] [Google Scholar]
- 37. Akbari E., Spychalski G. B., Rangharajan K. K., Prakash S., and Song J. W., Lab Chip 18(7), 1084–1093 (2018). 10.1039/C8LC00130H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis D. M., Mavrogiannis N., Gagnon Z., and Gerecht S., Biomicrofluidics 12(4), 042202 (2018). 10.1063/1.5026901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mannino R. G., Qiu Y., and Lam W. A., Biomicrofluidics 12(4), 042203 (2018). 10.1063/1.5024901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baratchi S., Tovar-Lopez F. J., Khoshmanesh K., Grace M. S., Darby W., Almazi J., Mitchell A., and McIntyre P., Biomicrofluidics 8(4), 044117 (2014). 10.1063/1.4893272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gossett D. R., Weaver W. M., MacH A. J., Hur S. C., Tse H. T. K., Lee W., Amini H., and Di Carlo D., Anal. Bioanal. Chem. 397(8), 3249–3267 (2010). 10.1007/s00216-010-3721-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Westein E., van der Meer A. D., Kuijpers M. J. E., Frimat J.-P., van den Berg A., and Heemskerk J. W. M., Proc. Natl. Acad. Sci. U. S. A. 110(4), 1357 (2013). 10.1073/pnas.1209905110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chang C. W., Cheng Y. J., Tu M., Chen Y. H., Peng C. C., Liao W. H., and Tung Y. C., Lab Chip 14(19), 3762–3772 (2014). 10.1039/C4LC00732H [DOI] [PubMed] [Google Scholar]
- 44. Sudarsan A. P. and Ugaz V. M., Proc. Natl. Acad. Sci. U. S. A. 103(19), 7228–7233 (2006). 10.1073/pnas.0507976103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garstecki P., Fuerstman M. J., Stone H. A., and Whitesides G. M., Lab Chip 6(3), 437–446 (2006). 10.1039/b510841a [DOI] [PubMed] [Google Scholar]
- 46. Gascoyne P. R. C. and Vykoukal J., Electrophoresis 23(13), 1973–1983 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khoshmanesh K., Almansouri A., Albloushi H., Yi P., Soffe R., and Kalantar-zadeh K., Sci. Rep. 5, 9942 (2015). 10.1038/srep09942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saffman P. G., J. Fluid Mech. 22(2), 385–400 (1965). 10.1017/S0022112065000824 [DOI] [Google Scholar]
- 49. Dudani J. S., Go D. E., Gossett D. R., Tan A. P., and Di Carlo D., Anal. Chem. 86(3), 1502–1510 (2014). 10.1021/ac402920m [DOI] [PubMed] [Google Scholar]
- 50. Sofela S., Sahloul S., Rafeie M., Kwon T., Han J., Warkiani M. E., and Song Y.-A., Lab Chip 18(4), 679–687 (2018). 10.1039/C7LC00998D [DOI] [PubMed] [Google Scholar]
- 51. Saggiomo V. and Velders A. H., Adv. Sci. 2(9), 1500125 (2015). 10.1002/advs.201500125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328(5986), 1662 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Satoh T., Sugiura S., Shin K., Onuki-Nagasaki R., Ishida S., Kikuchi K., Kakiki M., and Kanamori T., Lab Chip 18(1), 115–125 (2018). 10.1039/C7LC00952F [DOI] [PubMed] [Google Scholar]
- 54. Song J. W., Paek J., Park K.-T., Seo J., and Huh D., Biomicrofluidics 12(4), 042211 (2018). 10.1063/1.5027385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Macdonald N. P., Currivan S. A., Tedone L., and Paull B., Anal. Chem. 89(4), 2457–2463 (2017). 10.1021/acs.analchem.6b04546 [DOI] [PubMed] [Google Scholar]
- 56. Nguyen D., McLane J., Lew V., Pegan J., and Khine M., Biomicrofluidics 5(2), 022209 (2011). 10.1063/1.3576930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fan J., Men Y., Hao Tseng K., Ding Y., Ding Y., Villarreal F., Tan C., Li B., and Pan T., Biomicrofluidics 12(3), 034107 (2018). 10.1063/1.5030629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thurgood P., Baratchi S., Szydzik C., Mitchell A., and Khoshmanesh K., Lab Chip 17(14), 2517–2527 (2017). 10.1039/C7LC00350A [DOI] [PubMed] [Google Scholar]
- 59. Boyd-Moss M., Baratchi S., Di Venere M., and Khoshmanesh K., Lab Chip 16(17), 3177–3192 (2016). 10.1039/C6LC00712K [DOI] [PubMed] [Google Scholar]
- 60. Zhu J. Y., Thurgood P., Nguyen N., Ghorbani K., and Khoshmanesh K., Lab Chip 17(22), 3862–3873 (2017). 10.1039/C7LC00898H [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for detailed information of methods, additional experimental data, and videos showing the liquid-to-gel phase transition of petroleum jelly as well as operation of fabricated chips for the localised contracted channel, droplet generation, and particle lateral migration.