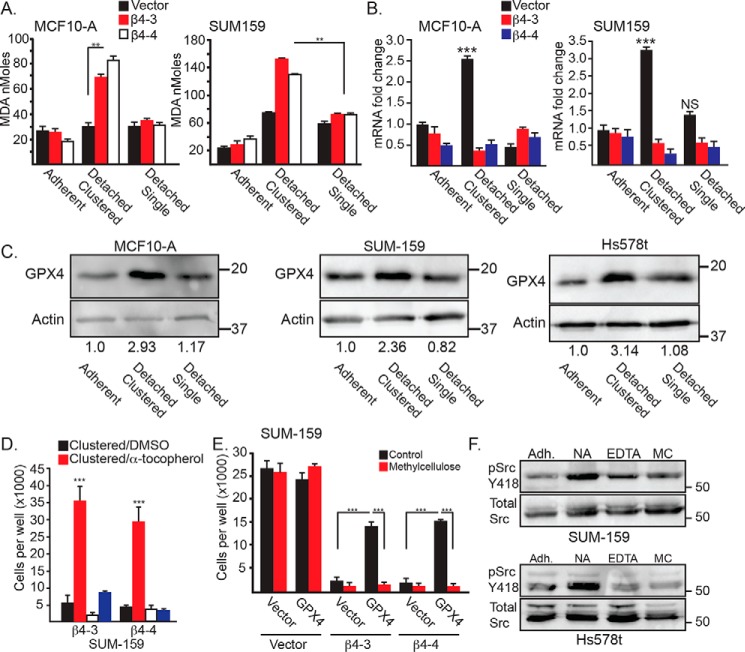
Figure 3.
Clustering of matrix-detached cells influences lipid peroxidation, GPX4 expression, and Src activity. A, lipid peroxidation was quantified using the MDA assay in control and β4-depleted MCF10-A and SUM-159 cells under either adherent, detached clustered (no additive), or detached single (0.5% methylcellulose) conditions for 4 h. Similar results were obtained using 2 mm EDTA. B, GPX4 mRNA expression was quantified by qPCR in control and β4-depleted MCF10-A and SUM-159 cells under either adherent, detached clustered (no additive) or detached single (0.5% methylcellulose) conditions for 2 h. C, GPX4 protein expression was assessed by immunoblotting in control MCF10-A, SUM-159, and Hs578t cells under detached clustered (no additive) or detached single (0.5% methylcellulose) conditions for 4 h. Immunoblots were quantified by densitometry, and the ratio of the intensity of the GPX4/actin bands relative to adherent cells is shown under the blots. D, SUM-159 β4-depleted cells were detached for 24 h with either no additive (clustered) or 2 mm EDTA (single) in the presence of either DMSO or 500 mm α-tocopherol, and the number of viable cells was quantified. E, control and β4-depleted cells SUM-159 cells that had been transfected with either a vector control or a GPX4 expression vector were detached for 24 h with no additive (Control) or 0.5% methylcellulose, and the number of viable cells was quantified. F, SUM-159 and Hs578t control cells were assessed for phosphorylated (Tyr-418) Src by immunoblotting under adherent conditions (Adh) as well as following 2 h of matrix detachment with no additive (NA), 2 mm EDTA (EDTA), or 0.5% methylcellulose (MC). NS, not significant. **, p < 0.01, ***, p < 0.005.