Figure 5.
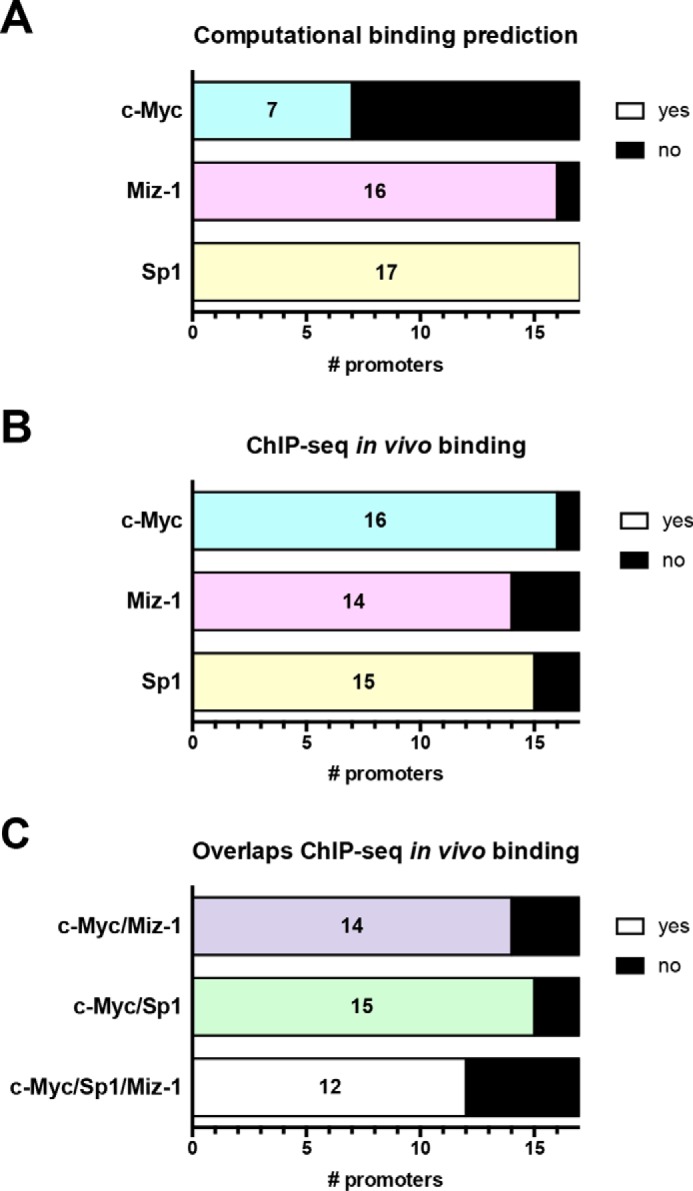
Computational binding predictions and in vivo binding data from ChIP-seq support an indirect model of repression by c-Myc through Sp1 and Miz-1. A, the promoters of miRNAs (for intergenic miRNAs) or their host genes (intronic miRNAs) (see Table S1) from the group of 19 were analyzed for computational binding prediction of c-Myc, Miz-1, and Sp1. B, using ChIP-seq data, in vivo binding for the three transcription factors was analyzed in all 17 promoters. C, the promoters were analyzed for overlaps between c-Myc and Miz-1, c-Myc, and Sp1 and c-Myc/Miz-1/Sp1 ChIP-seq data. A–C, the numbers indicate the number of promoters with a predicted binding site or overlapping ChIP-seq data.
