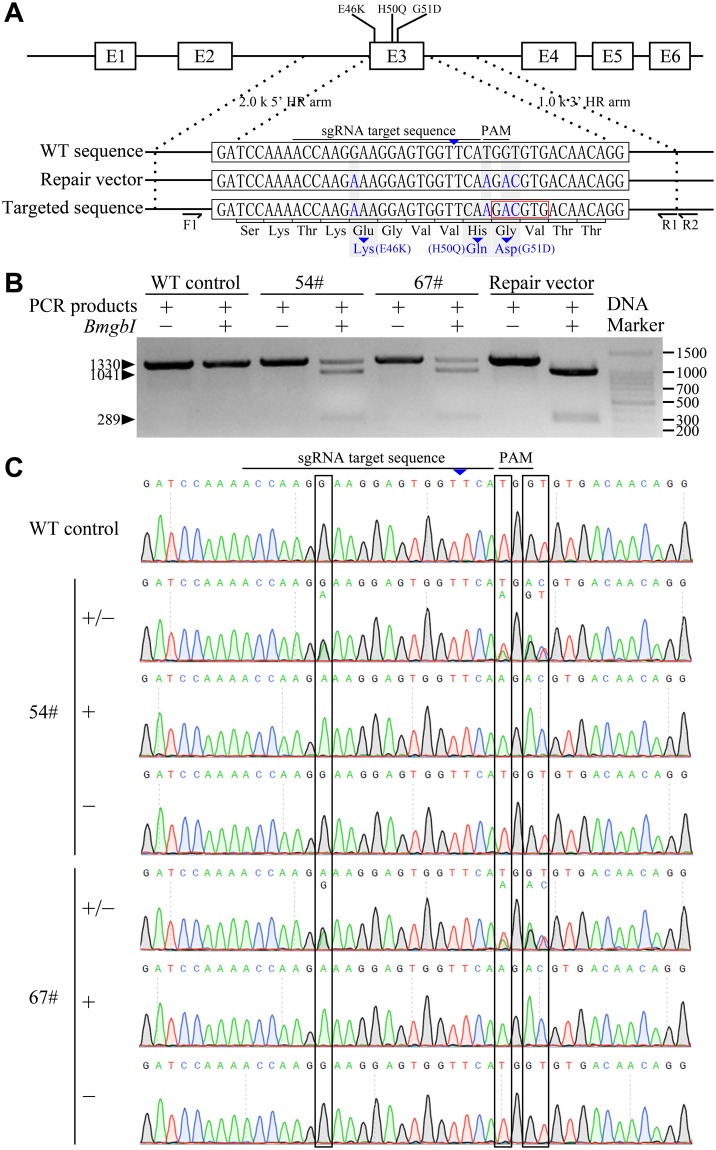
Figure 1.
Establishment of α-synuclein gene-edited Guangxi Bama minipig fibroblasts. (A) Strategy for precise editing of SCNA by CRISPR/Cas9-mediated homologous recombination (HR) with repair vector. The schematic presents the cleavage site on exon 3 of the SCNA locus, the three Parkinson’s disease-causing missense mutations (E46K, H50Q, and G51D), the small guide RNA (sgRNA) sequence, and the protospacer adjacent motif (PAM) sequence. Blue letters indicate the designed mutations on the SCNA locus (targeted sequence) and the corresponding mismatched nucleotides in the repair vector. The wild-type (WT) sequence of the SCNA locus is also shown. The BmgBI restriction enzyme site generated by the expected HR in the SCNA locus is denoted by a red box. (B) Cell colonies 54# and 67# carried monoallelic mutations in the SCNA locus and were identified by BmgBI digestion. WT cells were used as a negative control and the repair vector as a positive control. (C) DNA sequencing further confirmed the presence of the three desired mutations in cell colonies 54# and 67#. WT cells served as the negative control.