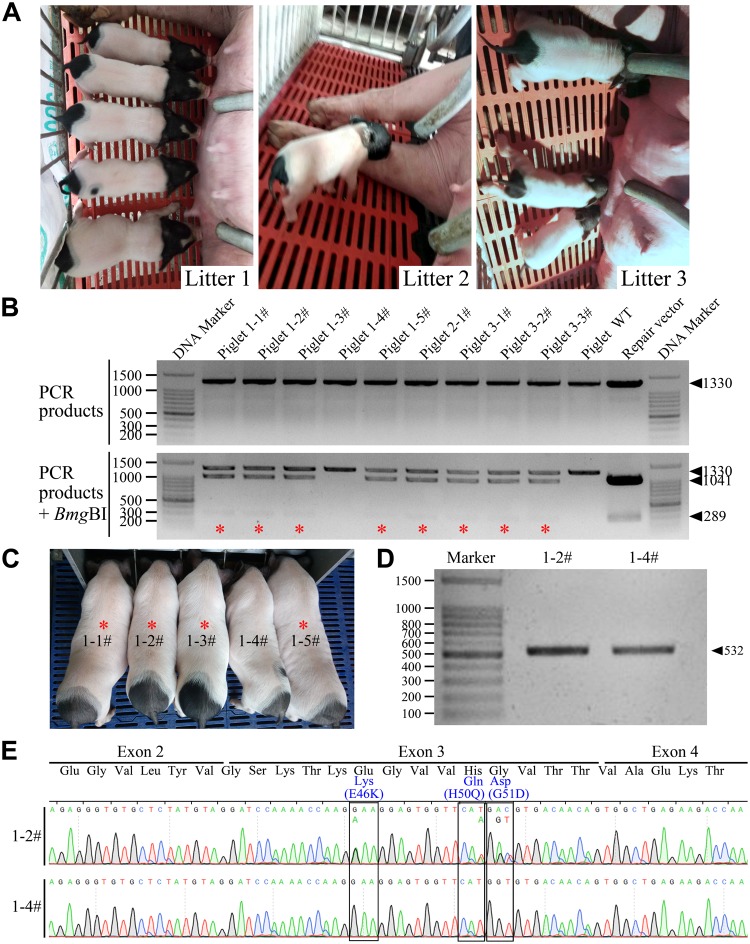
Figure 2.
Production and genotyping of gene-edited Guangxi Bama minipigs. (A) A total of nine cloned piglets from three litters appeared healthy at birth. (B) BmgBI digestion analysis showed that eight of nine piglets were monoallelic mutant at the SCNA locus (labeled by red asterisks), and the remaining one (1–4#) was a wild-type (WT). WT minipig were used as a negative control and the repair vector as a positive control. (C) Genotype-confirmed mutant minipigs (labeled by red asterisks) and the one wild-type minipig remained clinically healthy and showed normal growth and development at 3 months of age. (D) One mutant minipig (1–2#) and the age-matched wild type (1–4#) were used to confirm the SCNA mutations at the transcriptional level. The SCNA mRNA-coding sequence was obtained by RT-PCR. (E) The presence of the desired mutations at the transcriptional level (sequence coding for SCNA mRNA) was confirmed by DNA sequencing.