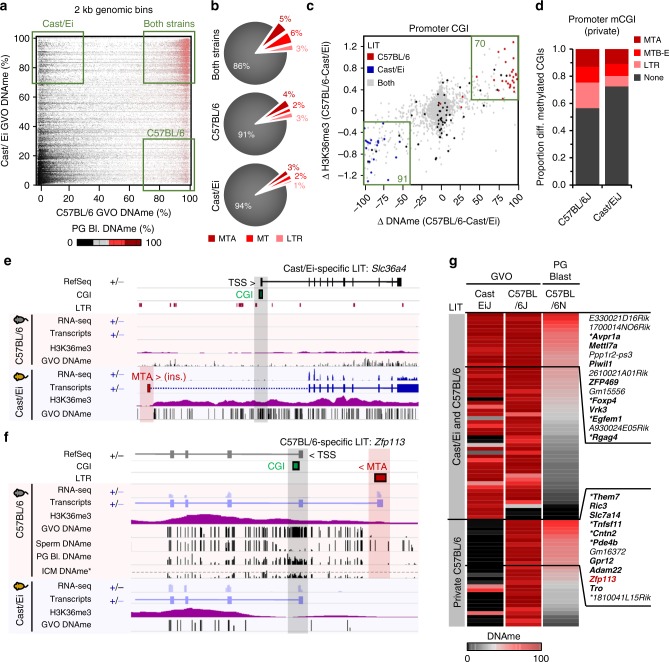
Fig. 6.
LTR polymorphisms lead to strain-specific transcripts and CGI DNAme in mouse oocytes. a Density plot of DNAme over 1,101,575 genome-wide 2 kb bins (>3 CpGs >1× coverage in Cast/Ei GVOs) in C57BL/6 or Cast/Ei GVOs. Regions hypermethylated (>70%) in C57BL/6 and/or Cast/Ei GVOs are highlighted. The mean percentage of DNAme in C57BL/6 parthenogenetic blastocysts (PG Bl.) is indicated as a color gradient. b Proportion of 2 kb bins hypermethylated in C57BL/6 and/or Cast/Ei GVOs overlapping with an LIT driven by an MTA element, another subfamily of MT element (MTB, C, D, or E) or another type of LTR. c Scatter plot of differential DNAme (Δ DNAme) and H3K36me3 enrichment (Δ H3K36me3) over 11,030 promoter CGIs in C57BL/6 and Cast/Ei GVOs. Differentially methylated CGIs are highlighted in green boxes. d Bar chart depicting each type of transcript overlapping differentially methylated promoter CGIs identified in c. MTA, MT, or LTR-initiated transcripts were identified by LIONs and/or manual inspection. None: no transcript or non-LTR initiated transcript. e Screenshot of the Slc36a4 CGI promoter. The Slc36a4 transcript initiates in a polymorphic MTA element (insertion in the Cast/Ei genome) in Cast/Ei GVOs. f Screenshot of the Zfp113 CGI promoter. The Zfp113 transcript initiates in an upstream MTA element in C57BL/6 GVOs but from the canonical TSS in Cast/Ei GVOs. g Heat map of all hypermethylated promoter CGIs (69) embedded within an LIT in C57BL/6 GVOs. DNAme levels in Cast/Ei GVOs, C57BL/6 GVOs, and C57BL/6 PG blastocysts is shown (columns), and CGIs embedded within LITs that are present in both strains or private to C57BL/6 oocytes are clustered (rows). Genes with CGI promoters retaining >45% DNAme in PG blastocysts are labeled. Bold: genes previously identified as TET targets at the implantation stage (see Fig. 5e). C57BL/6 GVO WGBS and ICM datasets from refs. 16,26