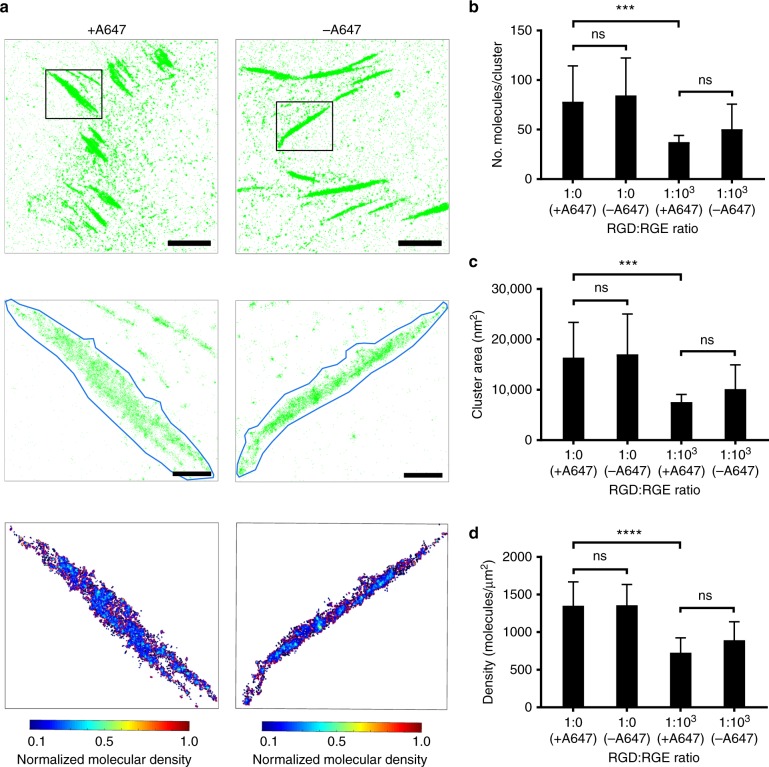
Fig. 3.
Paxillin-mEos2 clusters on RGD-A647- and RGD-functionalised ITO surfaces. a PALM images of paxillin-mEos2 in NIH-3T3 cells adhering onto ITO surfaces that were functionalised with RGD-A647 (left) and unlabelled RGD (right). Top: PALM images of paxillin-mEos2; scale bar = 5 µm. Middle: zoomed regions (black boxes in top row) of an individual adhesive structure with the region for analysis traced (blue line); scale bar = 1 µm. Bottom: paxillin-mEos2 clusters inside identified adhesive structures; colours indicated molecular densities. Images are representative images of n = 3−5 independent experiments. b Number of paxillin-mEos2 in clusters, c cluster area, and d density of paxillin-mEos2 molecules inside adhesive structures in NIH-3T3 cells on ITO surfaces with 1:0 and 1:103 of RGD-A647:RGE or RGD:RGE. Paxillin-mEos2 clustering inside adhesive structures was dependent on the RGD density on the surface, but not affected by A647 fluorophore labelled on RGD peptides; ns not significant (P > 0.05), ***P < 0.001 and ****P < 0.0001 (two-way ANOVA with Tukey post-testing). Bars and error bars are mean and standard deviation, respectively, n = 3−5 independent experiments (including 16−27 independent regions of interest)