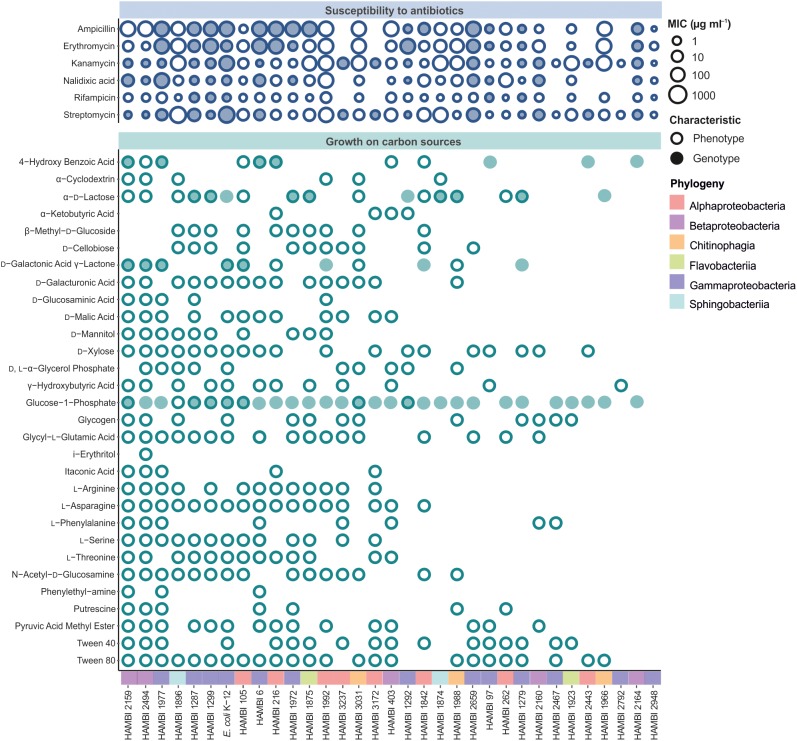
FIGURE 3.
Functional characteristics of model community strains. Both phenotypic (empty circle) and bioinformatically predicted (filled circle) susceptibility to antibiotics (upper panel) and ability to grow on a carbon source (lower panel) are shown. Matching between carbon source utilization phenotypes and functions predicted from genome sequences was only performed for 4-hydroxy benzoic acid, glucose-1-phosphate, D-galactonic acid γ-lactone, D-galacturonic acid and α-D-lactose where filled circles represent the presence of genomic modules for benzoic acid, glucose, galactonic acid, galacturonic acid and lactose metabolism, respectively. The strains are depicted in descending order of total number of carbon sources utilized. MIC, minimum inhibitory concentration.