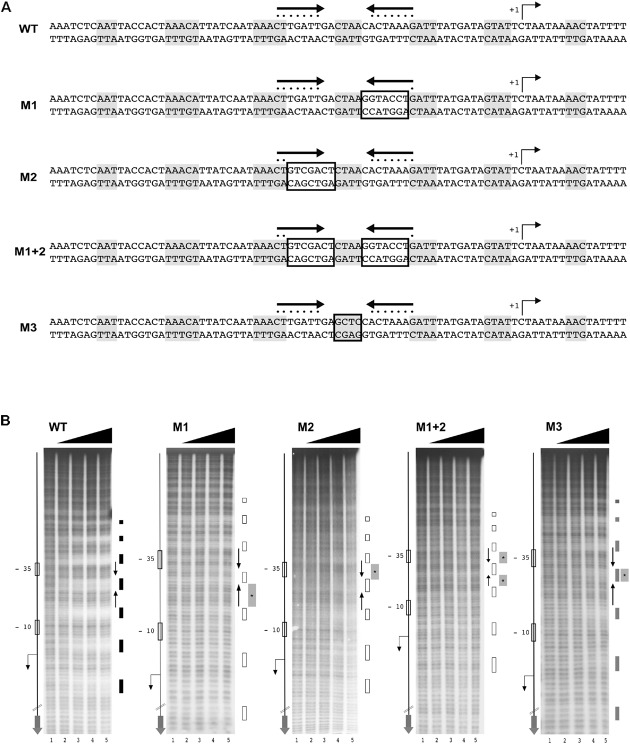
FIGURE 4.
The HAIR-like motif is essential for HspR binding to DNA. (A) Schematic representation of wild type and four different Pcbp mutant probes, in which one (M1 and M2) or both arms (M1 + 2) of the inverted repeat and the protected region between these sequences (M3) have been mutagenized by base substitution. HspR protected regions on Pcbp are shaded in gray and the inverted repeat sequences (HAIR-like motif) is represented by converging black arrows and their nucleotides marked with dots. In each DNA probe, mutagenized nucleotides are boxed. (B) From left to right, hydroxyl-radical footprint experiments on wild type and the indicated mutants of the Pcbp promoter region. Symbols are as described in the legend to Figure 3A. Wild type (WT) and indicated mutants DNA probes were incubated with increasing amounts of purified HspR protein and submitted to hydroxyl-radical digestion (see legend to Figure 3). Black, gray, and empty boxes to the right of each panel denote strong, weak, and loss of protection by HspR, respectively. Black converging arrows to the right of each panel mark the positions of the HAIR-like inverted repeat sequences, while gray boxes with an internal asterisk indicate the mutagenized regions.