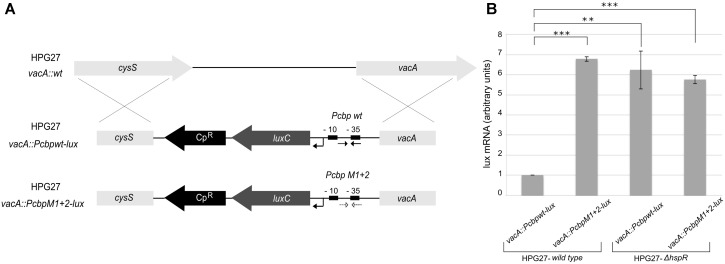
FIGURE 5.
In vivo H. pylori transcripts levels from the Pcbp promoter harboring WT or mutant HAIR-like sequences. (A) Schematic representation of Pcbpwt-lux and PcbpM1 + 2-lux reporter constructs obtained transforming the H. pylori G27 wild type acceptor strain by double homologous recombination in the vacA locus, and selected by chloramphenicol resistance (CpR). The wild type (Pcbpwt-lux) or HAIR-like mutant (PcbpM1 + 2-lux) Pcbp promoter is inserted upstream of a luxC reporter gene. The HAIR inverted repeat sequences are indicated by converging black arrows in the Pcbp WT promoter (Pcbpwt-lux) and by converging dotted arrows in the Pcbp HAIR-like mutant promoter (PcbpM1 + 2-lux). In each reporter construct, the –10 and –35 regions are depicted as black boxes and the transcriptional start site as a bent arrow. (B) Transcript levels of Pcbp wild type and Pcbp HAIR-like mutant promoters fused with lux reporter gene were assayed by qRT-PCR in the wild type and hspR deletion mutant strains using specific oligonucleotides for the luxC gene (LuxRTF/R). Mean values from three independent biological samples are reported in the graph, with error bars indicating standard deviation and asterisks marking statistical significance calculated by a Student’s t-test (∗∗∗p-value < 0.001; ∗∗p-value < 0.01).