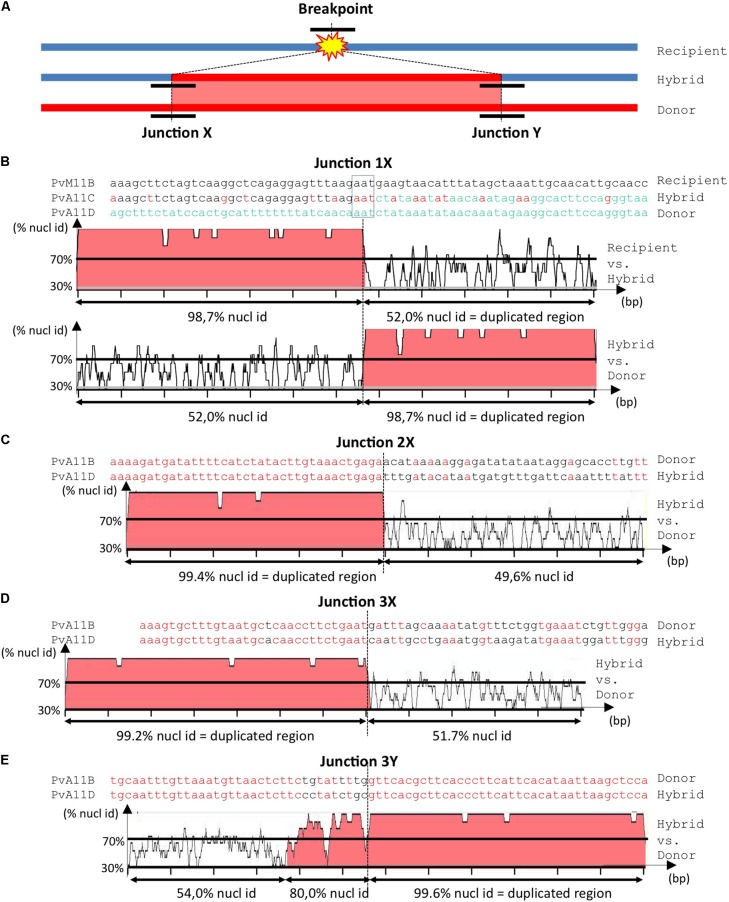
FIGURE 4.
Analysis of junctions from SD‘ events at Co-2 subtelomeric region of chromosome 11 long arm. (A) Simplified summary model describing the resulting stages of a SD event. The double-strand break occurs somewhere in the recipient (blue) molecule. Before repair, partial digestion of the breakpoint borders may occur, leading to loss of sequence. During the repair process, there is a duplicative invasion of the recipient molecule by the donor (red) molecule. Black short lines represent the regions that has to be aligned to accurately resolve the junctions (X and Y) between the broken molecule (recipient) and the duplicated region (donor), resulting in the final hybrid molecule. For more detailed information on double-strand break repair models, please refer to Puchta (2005) and Fiston-Lavier et al. (2007). (B–E) Nucleotide alignment at junctions 1X, 2X, 3X, and 3Y, respectively. Sequences are named according to their corresponding contigs. Aligned matching bases are red. For larger-scale vizualization, the 1000 bp surrounding region was analyzed using mVISTA (Mayor et al., 2000) after indels removal. Identity between sequences is indicated in %. Regions with identity equal or above 70% in 10bp windows are filled in red. Note that when the hybrid molecule is retained in the progeny, the recipient molecule is lost and a putative recipient molecule has to be found in another genotype to help resolving the breakpoint event. Therefore in (B) we used PvM11B contig from BAT93 genotype to help resolving junction 1X from an SD that occurred between PvA11B and PvA11C contigs in G19833 genotype. For junctions 2X (C), 3X (D) and 3Y (E), the recipient was not retrieved in BAT93, therefore the junctions are a bit less accurately resolved, but it was sufficient to infer the associated repair mechanism (i.e., NHEJ or HR).