Abstract
MicroRNA (miR)-200b-3p is downregulated in multiple human cancer types. Wnt signaling serves a role in human colorectal cancer (CRC). The present study aimed to examine the effect of miR-200b-3p on human CRC and its potential association with Wnt signaling. The Cell Counting Kit-8 (CCK-8) was employed to assess cell viability. A flow cytometric assay was conducted to examine cell proliferation and apoptosis. The regulation model of miR-200b-3p and Wnt1 was assessed by a luciferase reporter assay. A commercial kit was used to evaluate the activity of caspase-3 following treatment of the cells by miR-200b-3p or Wnt1. The expression of target factors was determined by a quantitative real-time polymerase chain reaction and western blot analysis. The expression of miR-200b-3p was decreased in human CRC tissues and in cell lines. The bioinformatics analysis and the luciferase reporter assay revealed that Wnt1 may be a direct target of miR-200b-3p. Moreover, the viability and proliferation of CRC cells was suppressed by miR-200b-3p. miR-200b-3p additionally induced apoptosis in CRC cells. Furthermore, the caspase-3 activity was enhanced in the miR-200b-3p mimics group. The expression of antigen Ki-67 (additionally termed KI-67) and β-catenin was decreased, while the expression of cleaved caspase-3 was increased by miR-200b-3p. In conclusion, miR-200b-3p inhibited proliferation and induced apoptosis in CRC cells by inactivating Wnt/β-catenin signaling. The present study provided potential biomarkers and candidate modalities for the management of CRC.
Keywords: miR-200b-3p, Wnt1, β-catenin, colorectal cancer
Introduction
Human colorectal cancer (CRC), one of the most common malignant tumors in China, is a leading cause of tumor-associated mortality worldwide (1–3). The occurrence of CRC is primarily due to age and lifestyle factors (4,5); however, it is additionally a consequence of a number of mutations and alterations in critical oncogenes and suppressor genes (6). Mutations most frequently occur in the Wnt signaling pathway (7). The Wnt signaling cascade is implicated in carcinogenesis (8), embryonic development (9), cell proliferation (10) and apoptosis (11). The canonical Wnt pathway is able to regulate gene transcription, in which β-catenin is involved (12). Thus, this pathway is additionally termed the Wnt/β-catenin signaling pathway. Through the binding of the Wnt protein to the N-terminal extracellular cysteine-rich domain of a Frizzled (Fz) family receptor, Wnt/β-catenin signaling begins and leads to the translocation of accumulated β-catenin into the nucleus, and eventually leads to the transcription of its target genes, including proto-oncogenes (12,13). It has been reported that mutation of β-catenin occurs in 10% of CRC cases (14). Accordingly, factors that may block the activity of Wnt signaling may be modalities for treating human CRC.
MicroRNAs (miRNAs), small non-coding RNAs, were recognized to have regulatory activity in the 2000s (15). Extensive evidence demonstrates that abnormal expression of miRNAs is a key step in tumor progression, in which process miRNAs serve as tumor suppressor genes or oncogenes. miRNAs are able to silence the expression of target mRNAs by binding to the complementary site in the 3′ untranslated region (3′ UTR) (16). It has been claimed that ~60% of genes in humans and other mammals are targets of miRNAs (17). The association of miRNAs and CRC was first identified in 2003 (18). Emerging evidence has demonstrated the role of miRNAs in regulating Wnt signaling, including in CRC (19). Wnt1, an important component of Wnt signaling, may serve as a target gene of miRNAs in multiple cell types (20–22). It has been reported that miRNA (miR)-200b-3p is a member of the miR-200 miRNA family, which is downregulated in a wide range of human cancer types including CRC (23,24). However, the association between miR-200b-3p and Wnt1 remains unclear.
Thus, the present study sought to examine the effect of miR-200b-3p on the proliferation and apoptosis of human CRC, and to clarify the potential associations between miR-200b-3p and Wnt signaling in the hopes it may provide clues to the mechanism of tumor progression in CRC.
Materials and methods
Tissue specimens
A total of 45 patients (male, mean age 58.8; range 37–75 years) with CRC were enrolled in the present study, with written informed consent. None of the patients received chemotherapy or radiotherapy prior to surgery at Yuhang Branch of Second Affiliated Hospital of Zhejiang University School of Medicine (Hangzhou, China) between April 2013 and June 2015. Patients who were histologically diagnosed with CRC and had complete clinicopathological data were included. Exclusion criteria included patients who had familial cancer syndrome and inflammatory bowel disease, a history of malignancy in other sites or infectious diseases. The CRC tissues and paracarcinoma tissues (≤5 cm distance from the tumor site) were obtained during the surgery. All the protocols in the present study were approved by the Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine.
Cell lines and cell culture
Human CRC cell lines (HCT-8, SW480, LOVO, HT29 and SW620) and a colonic epithelial cell line (NCM460) were purchased from the American Type Culture Collection (Manassas, VA, USA). The cells were maintained in RPM1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in the presence of 5% CO2.
Cell transfection and grouping
The cells were seeded at a density of 4×104 cells/well in a 24-well plate. When cells reached 50% confluence, the human miR-200b-3p mimics (10 nM; UAAUACUGCCUGGUAAUGAUGA cat. no. HMI0352; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), Wnt-1-small interfering (si)RNA (800 ng/µl; cat. no. HSH059152-CH1; GeneCopoeia, Inc., Rockville, MD, USA), si-negative control (800 ng/µl; cat. no. CSHCTR001-CH1; GeneCopoeia, Inc.) or miR-control (10 nM; cat. no. HMC0002; Sigma-Aldrich; Merck KGaA) was transfected using Lipofectamine® 2000 reagent (cat. no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. Following transfection for 48 h, the cells were collected for subsequent experiments. The experimental design was as follows: Control group (untreated cells, mock); negative control group (cells that were transfected with the siRNA negative control, NC); mimics group (cells that were transfected with miR-200b-3p mimics); and si-Wnt1 group (cells that were transfected with si-Wnt1). All the experiments were independently conducted at least three times.
Cell Counting Kit-8 (CCK-8) assay
The cells were seeded at a density of 1×103 cells/well in a 96-well plate following the aforementioned treatment. The cell viability was detected at different time points (12, 24 and 48 h post-siRNA transfection, respectively). CCK-8 solutions (10 µl) from the CCK-8 kit (cat. no. C0038; Beyotime Institute of Biotechnology, Haimen, China) were supplemented into the wells and incubated for 4 h. The absorbance of the reaction mixtures in each well was detected at 450 nm with a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
5(6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) assay
The cells (1×106 cells/ml) were stained with 5 µM CFSE (Invitrogen; Thermo Fisher Scientific, Inc.), and maintained for 10 min at 37°C. The labeling was stopped by adding medium with 10% FBS. Following a 5 min incubation on ice, the labeled cells were centrifuged at 700 × g at room temperature for 5 min, re-seeded in 24-well plate (1×105 cells/well) and treated as described previously in the present study. FACSCanto™ (BD Biosciences, Franklin Lakes, San Jose, CA, USA) was used to collect the CFSE fluorescence signals. The data were analyzed using FACSDiva software version 6.1.3 (BD Biosciences).
Flow cytometric analysis of apoptosis
The apoptosis of CRC cells was measured with the Annexin V-fluorescein isothiocycanate (FITC)/propidium iodide (PI) apoptosis detection kit (BioVision, Inc., Milpitas, CA, USA). The cells in every group were harvested and washed with PBS solution. The cells were incubated with Annexin V-FITC for 15 min in the dark at room temperature, and the incubation with PI was in the dark for 5 min at room temperature. The fluorescence intensity was analyzed using a flow cytometer (FACSCanto™, BD Biosciences). The data was analyzed with FlowJo software (FlowJo LLC).
Luciferase reporter assay
The bioinformatics analysis was performed using TargetScan (http://www.targetscan.org/vert_71/) and miRNA.org (http://34.236.212.39/microrna/home.do). The pLightSwitch-Wnt1-3′UTR marine RenSP luciferase reporter (cat. no. s812430) and Cypridina TK control vector (pTK-CLuc Vector) (cat. no. 32036) were purchased from Active Motif (Shanghai, China). The corresponding mutation vector was constructed as p-mut-Wnt1-3′UTR (CAGUAUU mutated to TACUCUC) using the MutBEST kit (Takara Bio, Inc., Otsu, Japan; cat. no. R401). The cells were plated in a 6-well plate (1×105 cells/well) and cultured overnight. Using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), the Cypridina TK control vector was co-transfected with pLightSwitch-Wnt1-3′UTR vector or p-mut-Wnt1-3′UTR vector into cells, respectively. Following 48 h, the luciferase activity was measured with a LightSwitch™ Dual Assay kit (Active Motif; cat. no. 32035) on a microplate reader (Bio-Rad Laboratories, Inc.). The Cypridina luciferase activity served as the control.
Caspase-3 activity
The cells in each group were harvested and re-suspended in lysis buffer provided by a caspase-3 assay kit (R&D systems, Inc., Minneapolis, MN, USA; cat. no. BF3100). Following centrifugation at 600 × g for 5 min at room temperature, the supernatant was mixed with reaction buffer and caspase-3 substrate in a 96-well plate, which was incubated for 2 h at 37°C. The absorbance at 405 nm was read with a microplate reader (Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract the total miRNA from tissues and cell lines, according to the manufacturer's protocol. The temperature protocol for the RT reaction was 25°C for 10 min, 42°C for 50 min and 70°C for 15 min. The concentration and the integrity were determined using a Nanodrop (Thermo Fisher Scientific, Inc.). cDNA was synthesized using 1 µg RNA with the PrimeScript RT Reagent kit (Takara Bio Inc.), according to the manufacturer's protocol. SYBR-Green PCR (Takara Bio Inc.) was used to perform the qPCR on the ABI 7500HT System (Thermo Fisher Scientific, Inc.). The primer pairs used were as follows: Wnt1 (forward), 5′-TGGCTGGGTTTCTGCTACG-3′; Wnt1 (reverse), 5′-CCCGGATTTTGGCGTATC-3′; miR-200b (forward), 5′-GCGGCTAATACTGCCTGGTAA-3′; miR-200b (reverse), 5′-GTGCAGGGTCCGAGGT-3′; Ki67 (forward), 5′-ACTGCAGCAGATGGAACTAGG-3′; Ki67 (reverse), 5′-AGAACAGTAGCGTGATGTTTGG-3′; β-catenin (forward), 5′-GCATGGGTCAGAAGGATTCCT-3′; β-catenin (reverse), 5′-TCGTCCCAGTTGGTGACGAT-3′; β-actin (forward), 5′-GCTGAGAACGGGAAGCTTGT-3′; β-actin (reverse), 5′-GCCAGGGGTGCTAAGCAG-3′; U6 (forward), 5′-CTCGCTTCGGCAGCACA-3′; and U6 (reverse), 5′-AACGCTTCACGAATTTGCGT-3′. The reactions were conducted using the following thermocycling conditions: Initial denaturation at 95°C for 5 min; 40 cycles of 95°C for 15 sec, and 60°C for 1 min; 72°C for 10 min for extension. The expression of target genes was calculated using the 2−ΔΔCq method (25). The expression fold change was relative to the internal housekeeping genes β-actin and U6.
Western blot analysis
Total protein from the cells was isolated using radioimmunoprecipitation assay buffer with protease inhibitors (Roche Diagnostics, Indianapolis, IN, USA). Proteins from tissue were isolated by a mechanical homogenizer for 5 min at 3,000 r/min. The concentrations of the proteins were quantified using a bicinchoninic acid kit (Beyotime Institute of Biotechnology). An equal amount of protein (25 µg) was loaded and separated by 8% SDS-PAGE. The proteins were electrotransferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). Bovine serum albumin (5%) (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) was added to block the non-specific antigens for 2 h at room temperature. The membranes were incubated with primary antibodies overnight at 4°C, and probed with secondary antibodies at room temperature for 1.5 h. The antibodies were as follows: Wnt1 (cat. no. sc-514531; 1:500; Santa Cruz Biotechnology, Inc.); β-catenin (cat. no. 2698; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA); Ki67 (cat. no. ab16667; 1:1,000 dilution; Abcam, Cambridge, UK); β-actin (cat. no. 3700; 1:1,000; Cell Signaling Technology, Inc.;); horseradish peroxidase-conjugated secondary antibody (cat. no. sc-2954; 1:200; Santa Cruz Biotechnology, Inc.). Following incubation, the membranes were washed with PBS containing 0.01% Tween-20. The bands were developed using enhanced chemiluminescence (ECL; Invitrogen; Thermo Fisher Scientific), following the manufacturer's protocol, with the LS45/55 chemiluminescence instrument (PerkinElmer, Inc., Waltham, MA, USA). In brief, the blot was incubated with working solution (provided by the ECL kit) by mixing equal parts of Detection Reagents 1 and 2 at room temperature for 1 min. Following removal of the working solution, the protected membranes in clear plastic wrap were placed in a film cassette, and subsequent X-ray exposure was performed. The intensity of the developed bands was calculated using Quantity One software version 4.6 (Bio-Rad Laboratories, Inc.).
Statistical analysis
GraphPad Prism version 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used to perform the data analysis. A two-tailed Student's t-test or one-way analysis of variance followed by Dunnett's multiple comparison test was performed to calculate the significant difference. Pearson's correlation coefficient (r) was used to measure correlation. P<0.05 was considered to indicate a statistically significant difference. The data are presented as the mean ± standard deviation. The experiments were repeated independently ≥3 times.
Results
Expression of miR-200b-3p and Wnt1 in CRC tissues
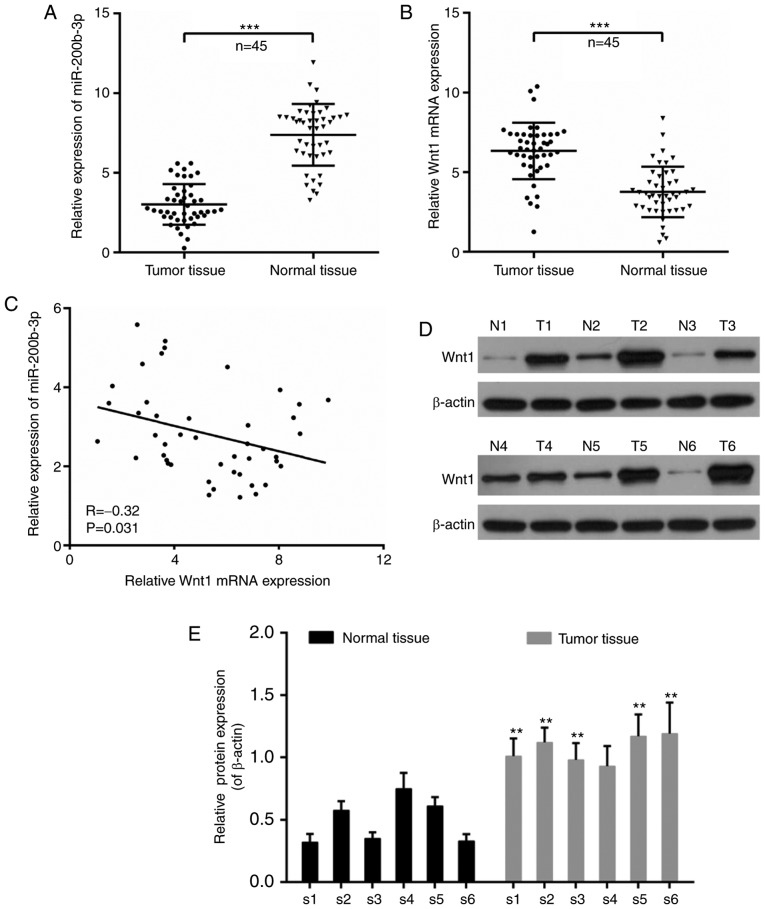
The dysregulation of miR-200b-3p and Wnt1 has been reported in tumorigenesis (26,27). Thus, the expression of miR-200b-3p and Wnt1 in patients with CRC was investigated. According to the RT-qPCR results from carcinoma tissues and paracarcinoma tissues, it is noteworthy that the expression of miR-200b-3p was significantly decreased in tumor tissues compared with that in paracarcinoma tissues (P<0.001; Fig. 1A). By contrast, the expression of Wnt1 was significantly increased in tumor tissues compared with that in normal tissues (P<0.001; Fig. 1B). Moreover, the expression of miR-200b-3p and Wnt1 was negatively correlated in CRC (r=−0.32; P=0.031; Fig. 1C). In addition, six cases were randomly selected to determine the expression of Wnt1 at the translational level. It was demonstrated that the expression of Wnt1 was increased in tumor tissues compared with that in normal tissues (Fig. 1D), with the exception that there was no significant difference in the fourth case (P<0.01; Fig. 1.E).
Figure 1.
Expression of miR-200b-3p and Wnt1 in CRC tissues. (A) qPCR for the expression of miR-200b-3p in CRC tissues and paracarcinoma tissues. (B) qPCR for the expression of Wnt1 in CRC tissues and adjacent normal tissues. (C) The expression correlation between miR-200b-3p and Wnt1 in CRC tissues. (D) Western blot analysis of the expression of Wnt1 in six randomly selected cases. N indicates adjacent normal tissues; T indicates tumor tissues. (E) Relative Wnt1 protein expression in normal tissues and tumor tissues in the six patients. β-actin was used as a loading control. n=45. The data was shown as mean ± standard deviation. **P<0.01, ***P<0.001 vs. normal tissues. CRC, colorectal cancer; qPCR, quantitative polymerase chain reaction; miR, microRNA.
Expression of miR-200b-3p and Wnt1 in CRC cell lines
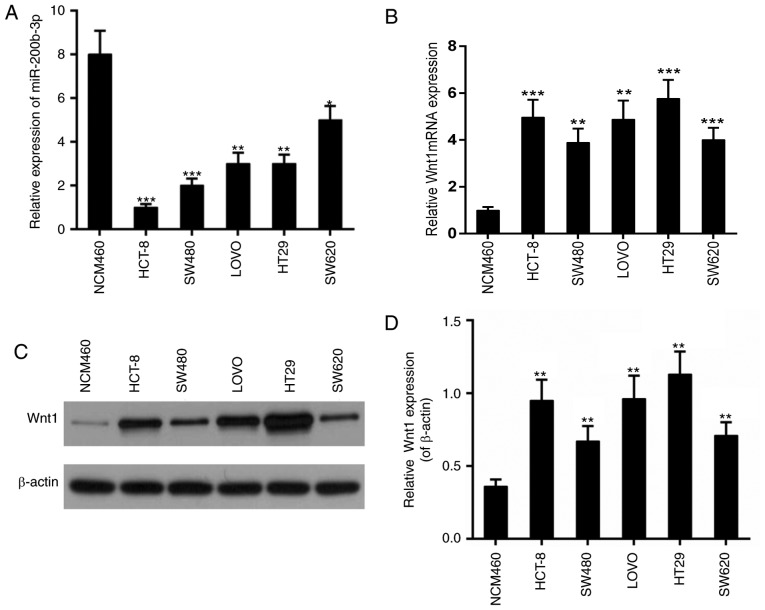
Subsequently, the expression of miR-200b-3p and Wnt1 was examined in different CRC lines including HCT-8, SW480, LOVO, HT29 and SW620. The colonic epithelial cell line NCM460 was used as a control. The results revealed that the expression of miR-200b-3p was decreased in all these CRC lines (Fig. 2A); whereas, the expression of Wnt1 was increased at the transcriptional and the translational level (Fig. 2B-D). The most marked alteration in the expression of miR-200b-3p and the expression of Wnt1 was observed in HCT-8. Thus, HCT-8 was selected for the subsequent experiments.
Figure 2.
Expression of miR-200b-3p and Wnt1 in CRC cell lines. (A) qPCR for the expression of miR-200b-3p in CRC cell lines. (B) qPCR for the expression of mRNA of Wnt1 in CRC cell lines. (C and D) Western blot analysis for the expression of Wnt1 in CRC cell lines. β-actin was used as a loading control. *P<0.05, **P<0.01, ***P<0.001 vs. NCM460 group. CRC, colorectal cancer; qPCR, quantitative polymerase chain reaction; miR, microRNA.
Wnt1 may be a direct target of miR-200b-3p
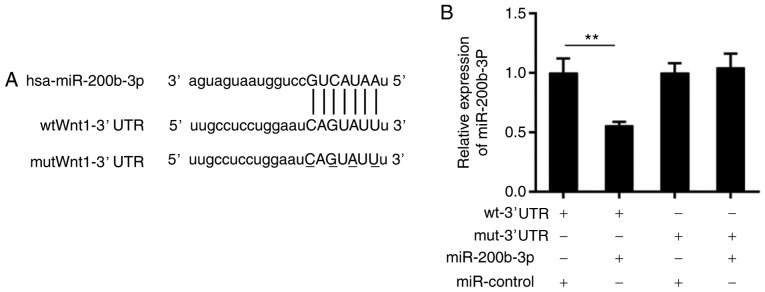
It is widely accepted that miRNAs exert function by silencing target mRNAs (28). Therefore, a bioinformatics analysis was performed using TargetScan (http://www.targetscan.org/vert_71/) and miRNA.org (http://34.236.212.39/microrna/home.do). The data from the two databases displayed that there was a potential binding site for miR-200b-3p in the 3′ UTR of the Wnt1 mRNA sequence (Fig. 3A). To verify this prediction, a luciferase reporter assay was conducted. It was revealed that the luciferase activity of wild-type Wnt1-3′UTR was significantly suppressed compared with the wild-type Wnt1-3′UTR and miR-control, by the presence of the miR-200b-3p mimics (P<0.01), which was not altered in the mutant Wnt1-3′UTR group (Fig. 3B).
Figure 3.
Wnt1 may be a direct target of miR-200b-3p. (A) The complementary sequences of miR-200b-3p in the Wnt1 mRNA 3′-UTR. (B) Luciferase activity of Wnt1 mRNA 3′-UTR (wt or mut) in the presence or the absence of miR-200b-3p by dual-luciferase reporter system. **P<0.01. UTR, untranslated region; wt, wild-type; mut, mutant-type; miR, microRNA.
Effect of miR-200b-3p on the cell viability, proliferation and apoptosis of CRC cells
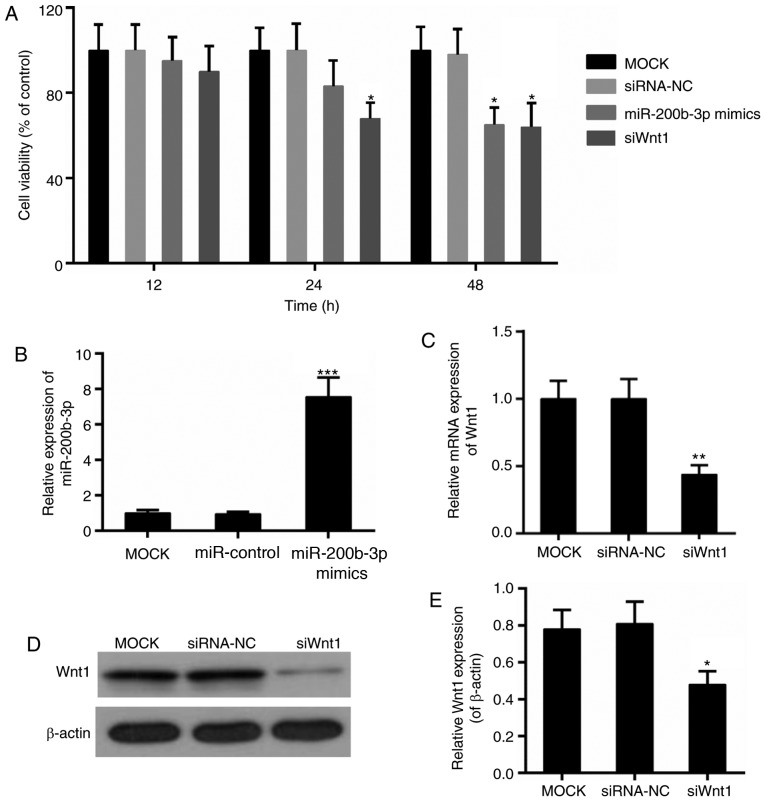
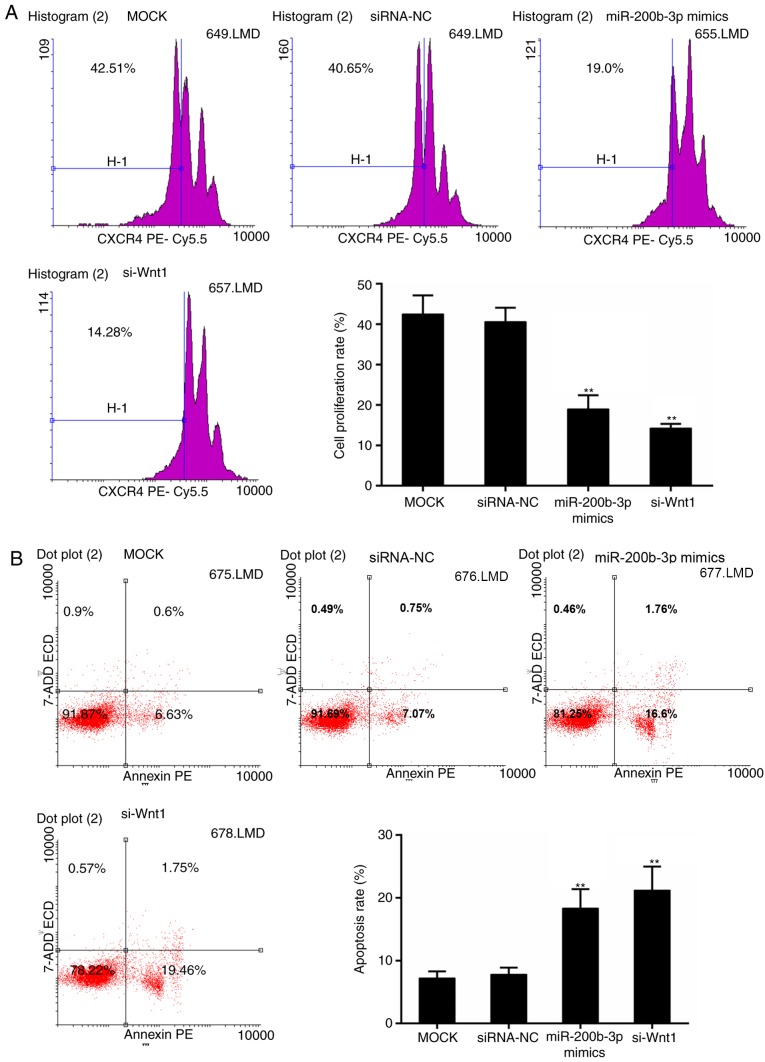
The effect of miR-200b-3p on CRC cells was detected with regards to cell viability, cell proliferation and apoptosis. The cell viability assay revealed that the Wnt1 interference suppressed the viability of CRC cells significantly, beginning at 24 h (P<0.05; Fig. 4A). By contrast, the significant decline in cell viability in the miR-200b-3p mimics group was observed following transfection for 48 h (P<0.05; Fig. 4A). In addition, the transfection efficiency of miR-200b-3p and Wnt1 was verified, respectively (Fig. 4B-E). Furthermore, the results from the CFSE assay revealed that cell proliferation was significantly inhibited in the miR-200b-3p mimics group and si-Wnt1 group compared with that in the control group, respectively (P<0.01; Fig. 5A). By contrast, the apoptosis level was significantly higher with the presence of miR-200b-3p mimics and si-Wnt1 compared with that in the control group, respectively (P<0.01; Fig. 5B).
Figure 4.
Effect of miR-200b-3p on the viability of CRC cells. (A) Cell viability detection by Cell Counting Kit-8 assay. (B) qPCR for the expression of miR-200b-3p following transfection. (C) qPCR for the expression of mRNA of Wnt1 following transfection. (D) Western blot analysis of the expression of Wnt1 following transfection. β-actin was used as a loading control. (E) qPCR for the expression of Wnt1 following transfection. *P<0.05, **P<0.01, ***P<0.001 vs. mock group. CRC, colorectal cancer; qPCR, quantitative polymerase chain reaction; siRNA-NC, small interfering RNA negative control; si-Wnt1, small interfering RNA-Wnt1; miR, microRNA.
Figure 5.
Effect of miR-200b-3p on the proliferation and apoptosis of CRC cells. (A) CFSE assay for cell proliferation. (B) Flow cytometric analysis for apoptosis. **P<0.01 vs. mock group. siRNA-NC, small interfering RNA negative control; si-Wnt1, small interfering RNA-Wnt1; CRC, colorectal cancer; CFSE, 5(6)-carboxyfluorescein diacetate succinimidyl ester; miR, microRNA.
Effect of miR-200b-3p on the expression of Ki67, cleaved caspase-3 and β-catenin
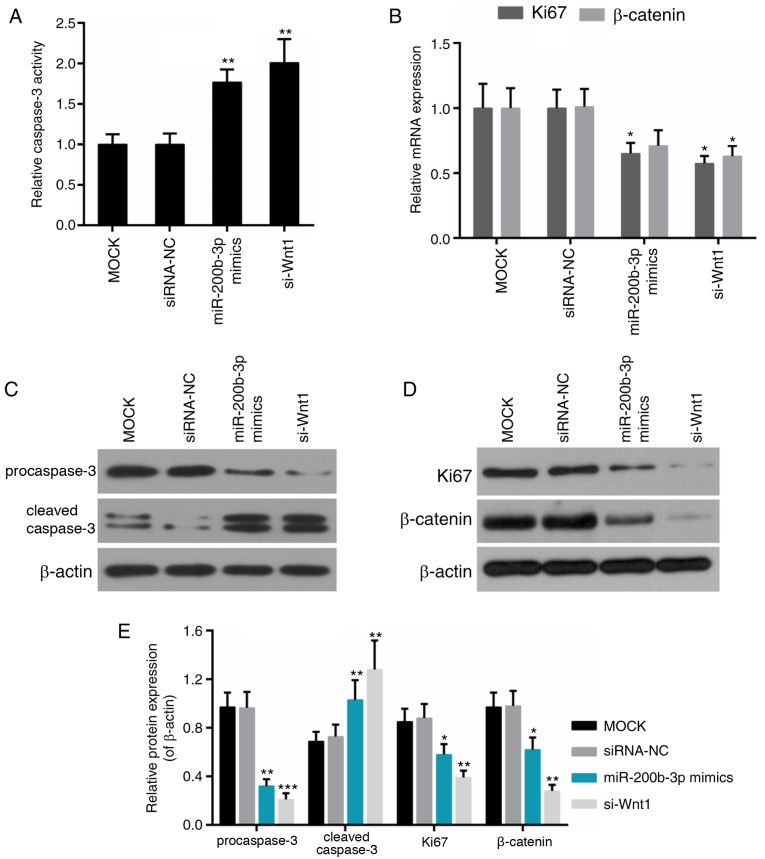
Caspase-3 is an important protein implicated in multiple apoptosis pathways (29), while Ki67 is recognized to be a cell proliferation-associated protein (30). As an important component of the canonical Wnt pathway, β-catenin is dysregulated in numerous carcinomas, including CRC (31); thus, the effect of miR-200b-3p on their expression was further examined. It was demonstrated that caspase-3 activity was elevated significantly in the miR-200b-3p mimics group and the si-Wnt1 group compared with that in the control group, respectively (P<0.01; Fig. 6A). Expression of Ki67 was reduced in the miR-200b-3p mimics group; the expression of Ki67 and β-catenin was decreased in si-Wnt1 group compared with that in the control group (Fig. 6B). Furthermore, the protein level of pro-caspase-3 and cleaved caspase-3 was decreased and increased in the miR-200b-3p mimics group and the si-Wnt1 group compared with that in the control group, respectively (Fig. 6C-E).
Figure 6.
Effect of miR-200b-3p on the activity of Ki67, β-catenin and caspase-3. (A) Relative caspase-3 activity. (B) qPCR for the expression of Ki67 and β-catenin. (C) Western blot analysis of the expression of pro caspase-3 and cleaved caspase-3. (D and E) Western blot analysis of the expression of Ki67 and β-catenin. β-actin was used as a loading control. *P<0.05, **P<0.01, ***P<0.001 vs. mock group. siRNA-NC, small interfering RNA negative control; si-Wnt1, small interfering RNA-Wnt1; miR, microRNA.
Discussion
The present study illustrated the role of miR-200b-3p in the proliferation and apoptosis of CRC, and highlighted the importance of increased expression of Wnt1 in CRC progression.
In the present study, miR-200b-3p was frequently decreased in the CRC tissues and cell lines, compared with the paracarcinoma tissues or normal cells. Decreased expression of miR-200b has been identified to be closely associated with poor survival in ovarian carcinomas (32). Another study also indicated that miR-200b may be a prognostic marker for breast cancer (33). These results indicated the potential of miR-200b as a valuable biomarker for tumors. Wnt1, as an important ligand involved in Wnt/β-catenin signaling (27), is generally upregulated in human cancer (34). Similarly, the expression of Wnt1 was elevated in the CRC tissues and cell lines in the present study. Furthermore, the protein expression of Wnt1 was increased in approximately all tumor tissues from patients with CRC. Additionally, the expression of miR-200b-3p and Wnt1 was negatively correlated. It was indicated that decreased miR-200b-3p and elevated Wnt1 expression may be critical in the progression of CRC.
In consideration of the regulatory machinery of miRNAs, it was proposed that the expression of Wnt1 may be downregulated by miR-200b-3p. Thus, bioinformatics analysis was undertaken to confirm this hypothesis. According to the data from available online databases, it was revealed that there was a complementary sequence of miR-200b-3p in the 3′UTR of Wnt1. Subsequently, a luciferase reporter assay was performed to confirm this prediction. The presented data revealed that the luciferase activity of wild-type Wnt1-3′UTR was reduced by miR-200b-3p mimics, whereas the activity of mutant Wnt1-3′UTR was not affected. It was indicated that Wnt1 may be a direct target of miR-200b-3p. Therefore, there was further speculation that miR-200b-3p may be a tumor suppressor of CRC by regulating Wnt signaling.
To confirm the effect of miR-200b-3p on CRC and the underlying mechanism, the proliferation and apoptosis behavior of CRC following miR-200b-3p transfection was examined. It was demonstrated that the viability of CRC cells and CSFE fluorescence intensity was decreased via miR-200b-3p. Cellular apoptosis in CRC was increased in the miR-200b-3p mimics group compared with the control group. A previous study reported that miR-200b-3p may inhibit the metastasis of prostate cancer (26). The data presented in the present study combined with previous results from another study demonstrated the tumor suppressive ability of miR-200b-3p (35).
Sequential activation of caspases is the hallmark event in cellular apoptosis. Caspase-3, as an executioner caspase, is at the convergence of multiple apoptosis pathways (29,36,37). Furthermore, Ki67, as a strictly cell proliferation-associated factor (30), is present in all active phases in cell cycle progression and is inactive in the G0 phase (38). The activity of these molecules, which are associated with apoptosis and cell proliferation, was investigated. The present data demonstrated that the activity of caspase-3 was enhanced by miR-200b-3p. The expression of Ki67 was deduced and cleaved caspase-3 was enhanced in the presence of miR-200b-3p mimics. Furthermore, the abnormal activation of Wnt/β-catenin signaling is frequently identified in CRC (39,40). Therefore, the effect of miR-200b-3p on the expression of β-catenin was examined in the present study. The protein expression level of β-catenin was decreased via miR-200b-3p, although its mRNA expression was reduced slightly. It was suggested that miR-200b-3p decreased the activity of β-catenin. In addition, the inactive status of Wnt signaling may lead to the degradation of β-catenin (41). Thus, the decreased β-catenin protein expression level may be in part due to the silencing of Wnt1 that was mediated through miR-200b-3p. Taken together, it was suggested that miR-200b-3p may suppress the tumor progression of CRC by inhibiting Wnt/β-catenin signaling. The present study proposed that Wnt1 and miR-200b-3p may serve as potential biomarkers for predicting the occurrence of CRC and candidate modalities for targeted therapy for CRC.
A limitation of the present study was that the direct binding of miR-200b-3p to Wnt1 requires validation by in-depth investigations. A previous study reported that histone deacetylase 1/4 and specificity protein 1 (Sp1) may regulate the miR-200b expression, thereby affecting chemoresistance and epithelial-mesenchymal transition, respectively (42,43). However, another previous study claimed that Sp1 was a target of miR-200b (33). Thus, future examination of the upstream regulator may be interesting and valuable for understanding the molecular mechanisms underlying the progression of CRC. In addition, the miR-200 family has been demonstrated to have an anti-tumor effect (44). Thus, determining the combined effect of miR-200b-3p and other family members on CRC may inspire another promising strategy for the treatment of CRC.
In conclusion, the present study uncovered valuable clues underpinning the mechanism of tumor progression in CRC. The present data revealed that miR-200b-3p and Wnt1 serve important roles in the progression of CRC cells. Wnt1 may be a direct target of miR-200b-3p. Furthermore, miR-200b-3p suppressed proliferation and induced apoptosis in CRC by regulating the activity of caspase-3, Ki67 and β-catenin. These results may have clinical implications for patients with CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Zhejiang Provincial Medical Science and Technology Project (grant no. 2017KY565).
Availability of data and materials
All data generated and/or analyzed during this study are included in this published article.
Authors' contributions
LC wrote the main manuscript. LC, XW, YZ and JZ performed the experiments. LC and QL designed the study. XW and YZ performed data analysis. All authors reviewed the manuscript.
Ethics approval and consent to participate
All the protocols in the present study were approved by the Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine and written informed consent was obtained.
Consent for publication
Informed consent was obtained from all participants for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Vaiopoulos AG, Athanasoula KC, Papavassiliou AG. Epigenetic modifications in colorectal cancer: Molecular insights and therapeutic challenges. Biochim Biophys Acta. 2014;1842:971–980. doi: 10.1016/j.bbadis.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Watson AJM, Collins PD. Colon cancer: A civilization disorder. Dig Dis. 2011;29:222–228. doi: 10.1159/000323926. [DOI] [PubMed] [Google Scholar]
- 5.Lee IM, Shiroma EJ, Lobelo F, Puska P, Blair SN, Katzmarzyk PT. Lancet Physical Activity Series Working Group: Effect of physical inactivity on major non-communicable diseases worldwide: An analysis of burden of disease and life expectancy. Lancet. 2012;380:219–229. doi: 10.1016/S0140-6736(12)61031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fearon ER, Pardoll DM, Itaya T, Golumbek P, Levitsky HI, Simons JW, Karasuyama H, Vogelstein B, Frost P. Interleukin-2 production by tumor cells bypasses T helper function in the generation of an antitumor response. Cell. 1990;60:397–403. doi: 10.1016/0092-8674(90)90591-2. [DOI] [PubMed] [Google Scholar]
- 7.Basu S, Haase G, Ben-Ze'ev A. Wnt signaling in cancer stem cells and colon cancer metastasis. F1000Res. 2016;19:5. doi: 10.12688/f1000research.7579.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giles RH, van Es JH, Clevers H. Caught up in a wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 9.Capdevila J, Belmonte lzpisúa JC. Extracellular modulation of the Hedgehog, Wnt and TGF-beta signalling pathways during embryonic development. Curr Opin Genet Dev. 1999;9:427–433. doi: 10.1016/S0959-437X(99)80065-3. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka S, Terada K, Nohno T. Canonical Wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. J Mol Signal. 2011;6:12. doi: 10.1186/1750-2187-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Guttridge DC, You Z, Zhang Z, Fribley A, Mayo MW, Kitajewski J, Wang CY. WNT-1 Signaling Inhibits Apoptosis by Activating β-Catenin/T Cell Factor–Mediated Transcription. J Cell Biol. 2001;152:87–96. doi: 10.1083/jcb.152.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 13.Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 14.Dai X, Wang L, Zhang L, Han Y, Yang G, Li L. The expression and mutation of beta-catenin in colorectal traditional serrated adenomas. Indian J Pathol Microbiol. 2012;55:288–293. doi: 10.4103/0377-4929.101732. [DOI] [PubMed] [Google Scholar]
- 15.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 16.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:E65. doi: 10.1186/gb-2004-5-9-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 19.Fan D, Lin X, Zhang F, Zhong W, Hu J, Chen Y, Cai Z, Zou Y, He X, Chen X, et al. MicroRNA 26b promotes colorectal cancer metastasis by down-regulating pten and wnt5a. Cancer Sci. 2017;109:354–362. doi: 10.1111/cas.13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood. 2009;114:404–414. doi: 10.1182/blood-2008-09-179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S, Zhang L, Yang P, Chen P, Xie Y. HCV core protein-induced down-regulation of microRNA-152 promoted aberrant proliferation by targeting Wnt1 in HepG2 cells. PLOS One. 2014;9:e81730. doi: 10.1371/journal.pone.0081730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si W, Li Y, Shao H, Hu R, Wang W, Zhang K, Yang Q. MiR-34a Inhibits Breast Cancer Proliferation and Progression by Targeting Wnt1 in Wnt/β-catenin Signaling Pathway. Am J Med Sci. 2016;352:191–199. doi: 10.1016/j.amjms.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 24.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Williams LV, Veliceasa D, Vinokour E, Volpert OV. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS One. 2013;8:e83991. doi: 10.1371/journal.pone.0083991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei W, Chua MS, Grepper S, So SK. Blockade of Wnt-1 signaling leads to anti-tumor effects in hepatocellular carcinoma cells. Mol Cancer. 2009;8:76. doi: 10.1186/1476-4598-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porter AG, Janicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 30.Scholzen T, Gerdes J. The Ki-67 protein: From the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Gao ZH, Lu C, Wang MX, Han Y, Guo LJ. Differential β-catenin expression levels are associated with morphological features and prognosis of colorectal cancer. Oncol Lett. 2014;8:2069–2076. doi: 10.3892/ol.2014.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leskela S, Leandro-Garcia LJ, Mendiola M, Barriuso J, Inglada-Perez L, Munoz I, Martinez-Delgado B, Redondo A, de Santiago J, Robledo M, et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr Relat Cancer. 2010;18:85–95. doi: 10.1677/ERC-10-0148. [DOI] [PubMed] [Google Scholar]
- 33.Yao Y, Hu J, Shen Z, Yao R, Liu S, Li Y, Cong H, Wang X, Qiu W, Yue L. MiR-200b expression in breast cancer: A prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med. 2015;19:760–769. doi: 10.1111/jcmm.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He B, You L, Uematsu K, Xu Z, Lee AY, Matsangou M, Mccormick F, Jablons DM. A monoclonal antibody against Wnt-1 induces apoptosis in human cancer cells. Neoplasia. 2004;6:7–14. doi: 10.1016/S1476-5586(04)80048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphries B, Yang C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472–6498. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghavami S, Hashemi M, Ande SR, Yeganeh B, Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ, Los M. Apoptosis and cancer: Mutations within caspase genes. J Med Genet. 2009;46:497–510. doi: 10.1136/jmg.2009.066944. [DOI] [PubMed] [Google Scholar]
- 37.Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Bruno S, Darzynkiewicz Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif. 1992;25:31–40. doi: 10.1111/j.1365-2184.1992.tb01435.x. [DOI] [PubMed] [Google Scholar]
- 39.Rafael S, Veganzones S, Vidaurreta M, de la Orden V, Maestro ML. Effect of β-catenin alterations in the prognosis of patients with sporadic colorectal cancer. J Cancer Res Ther. 2014;10:591–596. doi: 10.4103/0973-1482.139161. [DOI] [PubMed] [Google Scholar]
- 40.Jung YS, Jun S, Lee SH, Sharma A, Park JI. Wnt2 complements Wnt/β-catenin signaling in colorectal cancer. Oncotarget. 2015;6:37257–37268. doi: 10.18632/oncotarget.6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Chen DQ, Pan BZ, Huang JY, Zhang K, Cui SY, De W, Wang R, Chen LB. HDAC 1/4-mediated silencing of microRNA-200b promotes chemoresistance in human lung adenocarcinoma cells. Oncotarget. 2014;5:3333–3349. doi: 10.18632/oncotarget.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolesnikoff N, Attema JL, Roslan S, Bert AG, Schwarz QP, Gregory PA, Goodall GJ. Specificity protein 1 (Sp1) maintains basal epithelial expression of the miR-200 family: Implications for epithelial-mesenchymal transition. J Biol Chem. 2014;289:11194–11205. doi: 10.1074/jbc.M113.529172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manavalan TT, Teng Y, Litchfield LM, Muluhngwi P, Al-Rayyan N, Klinge CM. Reduced expression of miR-200 family members contributes to antiestrogen resistance in LY2 human breast cancer cells. PLOS One. 2013;8:e62334. doi: 10.1371/journal.pone.0062334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and/or analyzed during this study are included in this published article.