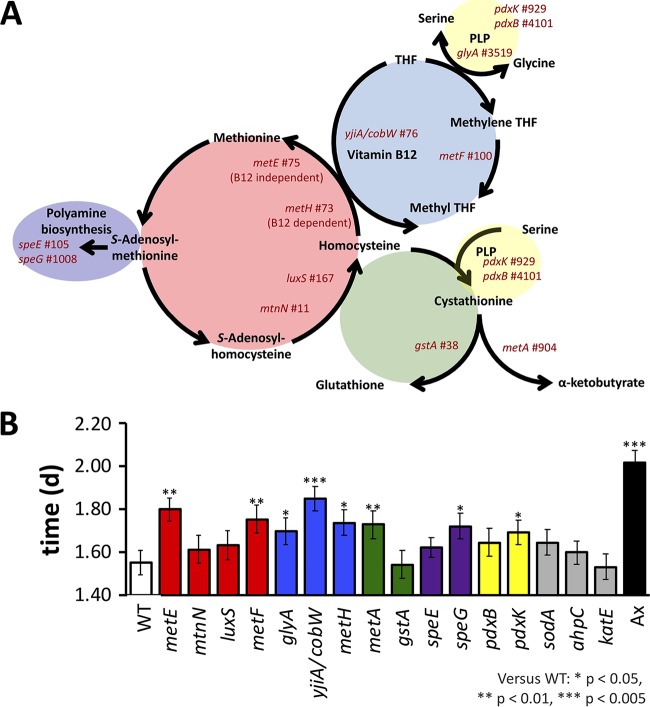
FIG 4.
Methionine cycle mutants decrease SR. SR of D. melanogaster that was monoassociated with E. coli methionine cycle mutants was compared to SR of D. melanogaster bearing a wild-type (WT) E. coli strain (4-letter codes from Table 2). (A) A simplified overview of the bacterial methionine biosynthesis pathway and related contributing pathways. Each mutant tested is listed near its pathway location, and mutants in the same subcycle are arranged by color. Gene name and MGWA PDG ranking (number to the right) are shown. The schematic is based on the work of Selhub (101). (B) Bacterial mutations that significantly influenced SR relative to the background E. coli control (WT) are indicated by asterisks. Ax, axenic. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (calculated by a Cox mixed-effects survival model). THF, tetrahydrofolate; PLP, pyridoxal 5′-phosphate.