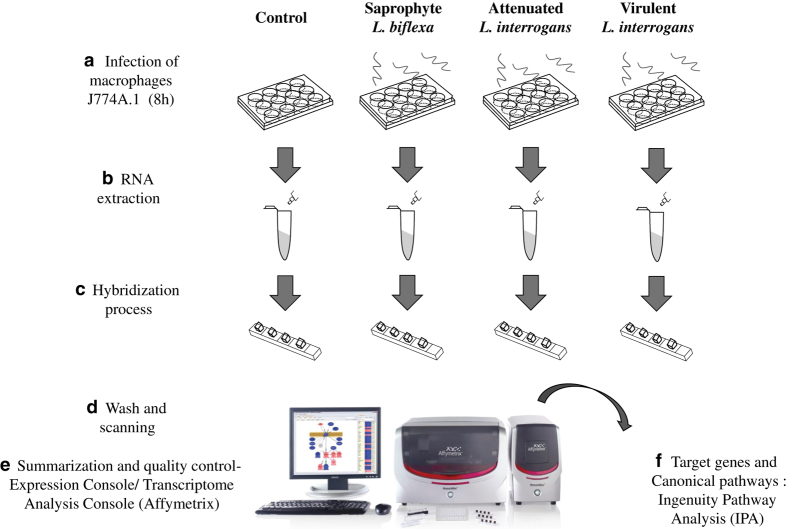
Figure 1. Illustration of experimental design.
Cell lineage J774A.1 of murine macrophages was cultured to a confluent monolayer. Infection was performed adding 100:1 bacteria:cell to the macrophages. (a) Treatments, analyzed in triplicate, were carried as follows: infection of macrophages with a virulent strain of L. interrogans, with an attenuated strain of L. interrogans, and with an saprophyte strain L. biflexa and non-infected macrophages as controls. All treatments were incubated in fresh RPMI medium, without antibiotics, for 8 h at 37 °C, 5% C02. (b) Following this period, total RNA was immediately extracted, (c,d) hybridization of samples to the strips was carried at 48 °C for 20 h, strips were then washed, stained and scanned using the GeneAtlas® System (Affymetrix). (e) Raw intensity values were background corrected, log2 transformed and then quantile normalized by the software Expression Console (Affymetrix) using the Robust Multi-array Average (RMA) algorithm. Statistical analysis was performed in the TAC software (Affymetrix) and cel files were submitted to Gene Expression Omnibus repository (GEO). (f) Target genes and Pathway analysis was performed in the Ingenuity Pathway Analysis (Qiagen).