Abstract
To study the evolution of drug resistance, we genetically and biochemically characterized Mycobacterium tuberculosis strains selected in vitro for ethambutol resistance. Mutations in decaprenylphosphoryl-β-d-arabinose (DPA) biosynthetic and utilization pathway genes Rv3806c, Rv3792, embB and embC accumulated to produce a wide range of ethambutol minimal inhibitory concentrations (MICs) that depended on mutation type and number. Rv3806c mutations increased DPA synthesis, causing MICs to double from 2 to 4 µg/ml in a wild-type background and to increase from 16 to 32 µg/ml in an embB codon 306 mutant background. Synonymous mutations in Rv3792 increased the expression of downstream embC, an ethambutol target, resulting in MICs of 8 µg/ml. Multistep selection was required for high-level resistance. Mutations in embC or very high embC expression were observed at the highest resistance level. In clinical isolates, Rv3806c mutations were associated with high-level resistance and had multiplicative effects with embB mutations on MICs. Ethambutol resistance is acquired through the acquisition of mutations that interact in complex ways to produce a range of MICs, from those falling below breakpoint values to ones representing high-level resistance.
Drug-resistant and multidrug-resistant (MDR) M. tuberculosis are emerging worldwide threats1,2. M. tuberculosis seems to develop resistance to the first-line drugs isoniazid, rifampin, pyrazinamide and ethambutol through the acquisition of SNPs or occasional small chromosomal deletions3,4. Clinical M. tuberculosis strains are usually classified as drug susceptible or drug resistant, although it is well recognized that there is a range of MICs among resistant strains. Given this view, it is not surprising that most of the mutations conferring drug resistance discovered so far appear to cause resistance to individual drugs in a single step, and drug resistance in clinical diagnosis with M. tuberculosis has been largely treated as a dichotomous variable. Little attention has been paid to the evolutionary events that might occur before M. tuberculosis develop an MIC in the range associated with resistance or after a MIC corresponding with drug resistance has been achieved. Yet, stepwise increases in MICs for individual drugs have been described in Mycobacterium smegmatis5. Furthermore, one laboratory study showed that M. tuberculosis required two-step selection to reach high-level fluoroquinolone resistance6. Therefore, it is possible that the evolution of resistance is more complex than commonly assumed in M. tuberculosis, although the genetic steps involved in this process are currently unknown.
Resistance to the first-line tuberculosis drug ethambutol seems to be particularly likely to occur as part of a multistep process5,7,8. Both epidemiological and laboratory evidence suggest that mutations in the embCAB operon are responsible for resistance to ethambutol, especially the ‘canonical’ mutations in codon 306, 406 or 497 of embB5,7–10. The embCAB operon encodes arabinofuranosyltransferases that are involved in polymerizing arabinofuranosyl (Araf) residues from DPA into the arabinan components of cell wall arabinogalactan and lipoarabinomannan. The EmbB and EmbA enzymes transfer Araf residues to a galactan chain after the initial Araf residues are linked to galactan by another priming arabinofuranosyltransferase, AftA (encoded by Rv3792)11. EmbC uses the same donor DPA to transfer Araf residues to a mannan core after the initial Araf residues are linked to mannan by an uncharacterized arabinofuranosyltransferase12. DPA is synthesized in four steps from phosphoribosyl diphosphate (pRpp) and decaprenyl phosphate13 (Supplementary Fig. 1).
Despite a strong association between embB mutations and ethambutol resistance, many clinical strains have mutations in this gene although remaining susceptible to ethambutol14. We demonstrated that mutations in embB codons 306, 406 and 497 all caused ethambutol resistance but only at a low level of ethambutol between 6 and 14 µg/ml, depending on the specific mutation7,8. None of these mutations were found to produce the high-level resistance to ethambutol observed in clinical isolates. We considered the possibility that M. tuberculosis might develop a pre-resistant state characterized by ethambutol MICs below the critical threshold for drug resistance upon exposure to ethambutol. Such pre-resistant strains might be predisposed to evolve into more resistant organisms in a stepwise manner. By investigating the biological events that occur as M. tuberculosis evolves to high-level ethambutol resistance, our study identifies several mechanisms for the evolution of resistance to ethambutol, establishes the first known cause of high-level ethambutol resistance to our knowledge and provides more general insights into the evolution of M. tuberculosis drug resistance.
RESULTS
Mutations in Rv3806c are associated with small increases in ethambutol MICs, varying with genetic background
Canonical mutations in embB codons 306, 406 and 497 and non-canonical mutations in embB and in embC only explain approximately 60% of clinical ethambutol resistance. We isolated M. tuberculosis colonies with low-level resistance to ethambutol to identify additional resistance targets by plating ethambutol-susceptible M. tuberculosis strain 210 on medium with varying concentrations of ethambutol. Seven of the resulting colonies were found to have canonical mutations in embB (principally at codons 306 and 497)7,8. An additional five colonies contained wild-type embCAB sequences. We also identified five colonies that had a single embB mutation encoding p.Gln445Arg within the embCAB genes. Although each of these five isolates had an identical embB codon 445 mutation, their ethambutol MICs varied from 2 to 8 µg/ml in liquid medium.
We sequenced the entire genome of three isolates with wild-type embCAB (8C1, 8C6 and 16C10) and of one isolate (32C4) with a known embB codon 445 mutation and an ethambutol MIC of 4 µg/ml. We also sequenced two other embB codon 445 mutants (16C14 and 32C1) with ethambutol MICs of 8 and 2 µg/ml, respectively, at all of the sites found to be mutant in the four sequenced genomes. As controls, we resequenced the parental 210 strain and strain NJT210GTG, an ethambutol-resistant isogenic mutant of strain 210 that contains a SNP at embB codon 3067. The mutations present in each strain and how mutations potentially interacted to increase ethambutol MICs are shown in Table 1. Strain 8C1 had two synonymous mutations, one in Rv1251c and one in Rv2090, and two nonsynonymous mutations, one in rubB and one in Rv3806c. Strain 8C6 had one noncoding mutation and three nonsynonymous mutations in ripA, pepN and Rv0713. All strains had ethambutol MICs identical to that of the wild-type parental 210 strain as determined by liquid BACTEC testing but showed small MIC increases when tested by agar proportion. The embB codon 445 mutant 32C4 did not have any additional mutations outside of embB. However, the embB codon 445 mutant 16C14, which had the highest level of ethambutol resistance, contained an additional SNP in Rv3806c that was identical to the one found in strain 8C1.
Table 1.
Mutations generated by single-step selection for ethambutol resistance
| Strain | 210 (wild type) |
NJT210GTG (mutant control) |
8C1 | 8C6 | 16C10 | 16C14 | 32C1 | 32C4 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Ethambutol MIC (µg/ml) |
BACTEC 460TBa | 2 | 14 | 2 | 2 | 2 | 8 | 2 | 4 | |
| 7H10 agar proportiona | 2 | 14 | 6 | 4 | 6 | 16 | 6 | 6 | ||
| Location of mutationb | ||||||||||
| Rv number | Gene | Encoded alteration | ||||||||
| Rv713 | Conserved transmembrane | p.Thr237Met | −c | − | − | +d | − | − | − | − |
| Rv1251c | Conserved hypothetical | p.Leu1030Leu | − | − | + | − | − | − | − | − |
| Rv1477 | ripA | p.His400Asp | − | − | − | + | − | − | − | − |
| Rv2090 | Probable 5′–3′ exonuclease | p.Arg154Arg | − | − | + | − | − | − | − | − |
| Rv2467 | pepN | p.Arg41His | − | − | − | + | − | − | − | − |
| Rv3213c-gpm2 | Intergenic | T>G 46 bp upstream of Rv3213c; 108 bp upstream of gpm2 |
− | − | − | + | − | − | − | − |
| Rv3250c | rubB | p.Glu53Gly | − | − | + | − | − | − | − | − |
| Rv3795 | embB | p.Met306Val | − | + | − | − | − | − | − | − |
| p.Gln445Arg | − | − | − | − | − | + | + | + | ||
| Rv3806c | ubiA | p.Gly165Cys | − | − | + | − | − | + | − | − |
Method used to test MIC.
Gene sequence is wild type except where indicated.
−, mutation absent.
+, mutation present.
The Rv3806c gene encodes a 5-phospho-α-\FS18 d-ribose-1-diphosphate:decaprenyl-phosphate 5-phosphoribosyltransferase (DPPR synthase, UbiA)15,16. This enzyme synthesizes decaprenylphosphoryl-β-d-5-phosphoribose (DPPR), which is a precursor of DPA, the donor substrate for arabinosyltransferases, including EmbB and EmbC (Supplementary Fig. 1). We reasoned that Rv3806c might have a previously unknown role in ethambutol resistance, as Emb proteins are the presumed targets of ethambutol5. To further confirm the association between Rv3806c mutations and ethambutol resistance, we sequenced Rv3806c in an additional ten colonies that had been selected for growth in ethambutol but did not have any of the canonical embB mutations8. One of these isolates, 8C2, had a new SNP in Rv3806c causing a p.Leu235Pro substitution. Strain 8C2 had a wild-type embB gene and an ethambutol MIC of 4 µg/ml. These studies suggested that mutations in embB codon 445 and in Rv3806c are unlikely to have substantial effects on the ethambutol MIC when occurring alone but the ethambutol MIC may increase substantially when both embB codon 445 and Rv3806c mutations occur in the same genome.
Mutations in Rv3806c combined with mutations in embB and embC produce high-level ethambutol resistance
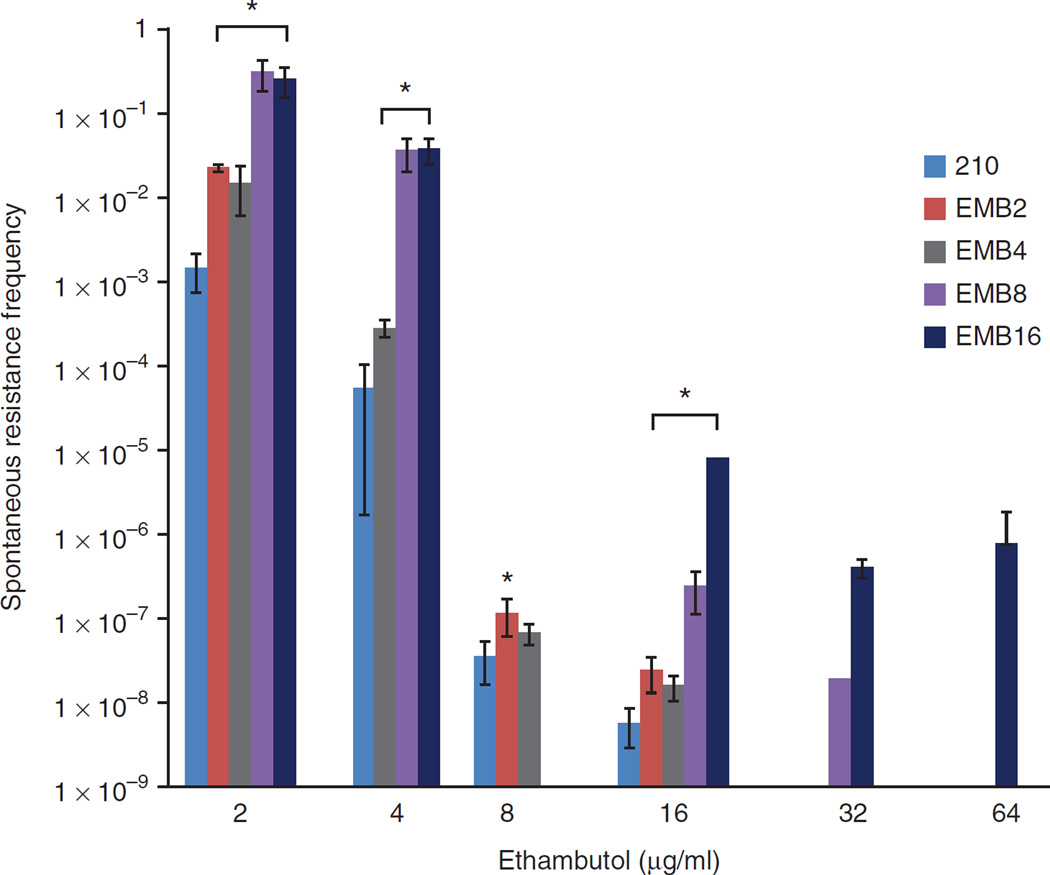
To determine whether high-level ethambutol resistance in M. tuberculosis can occur in a single step or whether a multistep process is required, we plated the 210 strain on medium containing 2, 4, 8, 16, 32 or 64 µg/ml ethambutol. Numerous colonies were obtained on the plates containing up to 16 µg/ml ethambutol but no growth was detected at higher ethambutol concentrations. Of the recoverable colonies, none tested had an ethambutol MIC of >16 µg/ml (Supplementary Table 1). In contrast, cultures taken from colonies that had grown on the plates with an ethambutol concentration of 8 or 16 µg/ml could easily be recovered when plated on plates with an ethambutol concentration of either 32 or 64 µg/ml, suggesting that high-level ethambutol resistance requires the occurrence of several mutations (Fig. 1).
Figure 1.
Frequencies of spontaneous resistance to ethambutol. Triplicate cultures of wild-type strain 210 and triplicate cultures of spontaneously ethambutol-resistant 210 mutants that had previously been selected with the indicated concentrations of ethambutol were replated on medium with ethambutol at the indicated concentrations. The mean frequency of colony-forming units (CFUs) recovered with each ethambutol concentration compared to in the no-drug control is shown. The number in each EMB ID indicates the ethambutol concentration (in µg/ml) under which in vitro strains were preselected. Error bars, s.d. of three independent experiments. *P < 0.05; significant differences compared to the 210 strain were determined by Student’s t test.
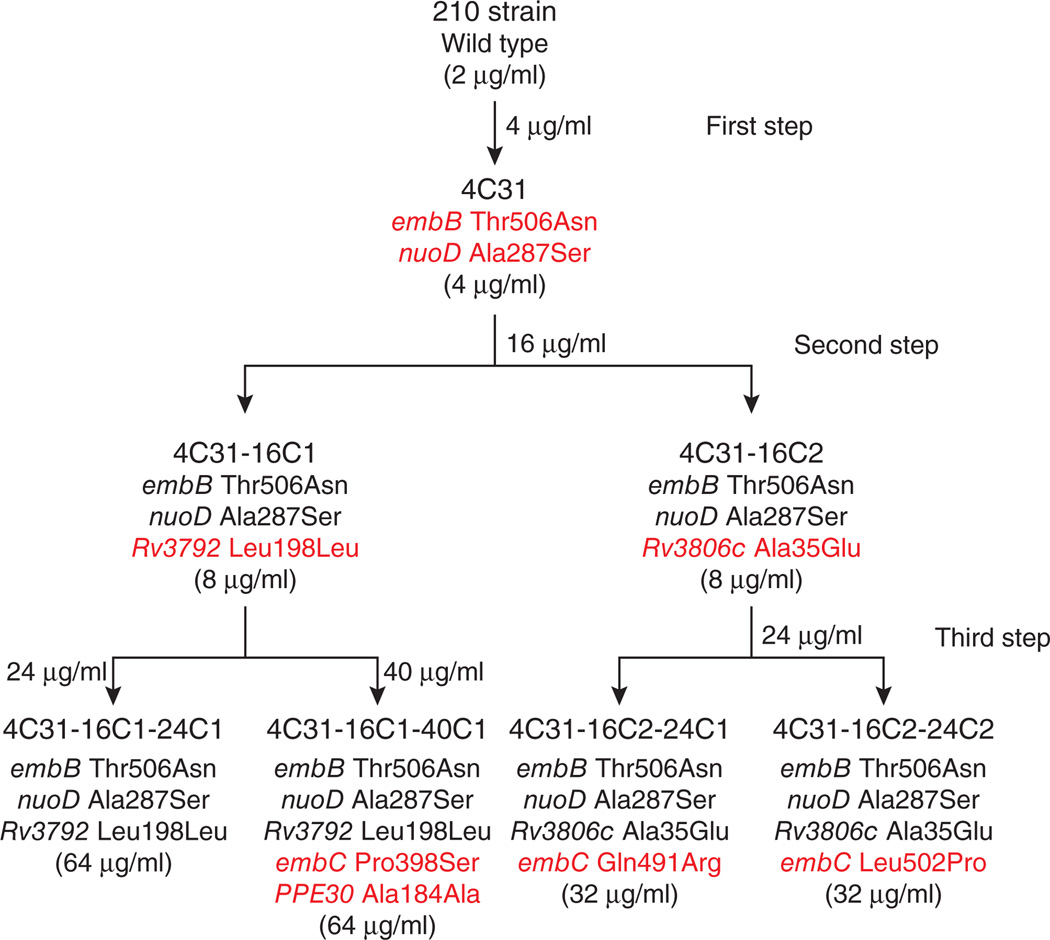
We further investigated one M. tuberculosis lineage that had undergone multistep selection for high-level ethambutol resistance through successive culturing of the ethambutol-susceptible 210 strain on plates with ethambutol concentrations of 4 and 16 µg/ml and then on plates with either 24 or 40 µg/ml ethambutol (Fig. 2). Each colony in the lineage was tested for its ethambutol MIC, and its whole genome was sequenced. In all but one case, each incremental increase in MIC was accompanied by at least one new mutation (confirmed by direct Sanger sequencing). In the first step, one mutation in embB (encoding a p.Thr506Asn substitution) and a second mutation in nuoD (encoding a p.Ala287Ser substitution) occurred as the strain increased its ethambutol MIC from 2 to 4 µg/ml. In the second step, both colonies increased their ethambutol MICs to 8 µg/ml. One colony acquired a single new mutation in Rv3806c encoding a p.Ala35Glu substitution. The second colony acquired a single SNP in Rv3792 leading to a synonymous p.Leu198Leu substitution. Finally, in the third step, four colonies showed very high ethambutol MICs ranging from 32 to 64 µg/ml. One colony had no new mutations, whereas the three other colonies each had one of the following new alterations encoded in embC: p.Pro398Ser, p.Gln491Arg or p.Leu502Pro. One colony had also developed an additional synonymous mutation in the PPE30 gene encoding p.Ala184Ala (Fig. 2). These results further suggest that high-level ethambutol resistance develops in a multistep process. In our lineage, the M. tuberculosis pathway toward high-level ethambutol resistance involved a mutation in embB in the first step, a mutation in either the DPA biosynthetic (Rv3806c) or utilization (Rv3792) pathway in the second step and a mutation in embC in the third step.
Figure 2.
Mutant strains and MICs generated during stepwise selection for ethambutol resistance. The phylogeny of the strains selected in vitro for multistep ethambutol resistance is shown. The ethambutol concentration used for selection is shown at each step. For each colony analyzed, shown (top to bottom) is the name assigned to the strain, the encoded alterations identified by whole-genome sequencing and the ethambutol MIC (shown in parentheses). Encoded alterations are shown in red in the generation in which they appear.
Rv3806c mutations are associated with high-level ethambutol resistance in clinical M. tuberculosis strains
We investigated the association between mutations in embB, Rv3806c, Rv3792, embC and nuoD and high-level ethambutol resistance in clinical M. tuberculosis. We sequenced these 5 genes in 63 clinical isolates randomly selected from the World Health Organization Special Programme for Research and Training in Tropical Disease strain bank to represent a broad range of ethambutol MICs as well as geographic and phylogenetic diversity17 (Supplementary Table 2). We identified embB, Rv3806c, Rv3792, embC and nuoD mutations in 48, 19, 12, 62 and 4 of these isolates, respectively. Sixty-two of the 63 clinical isolates also had a synonymous mutation in in embC encoding p.Arg927Arg, suggesting that this alteration was actually a marker for the H37Rv strain and closely related lineages. After eliminating this and other synonymous mutations and mutations that were strictly associated with strain lineage that could be considered phylogenetic markers, we found that mutations in embB codons 306, 354, 406 and 497 were significantly associated with ethambutol resistance (Table 2), confirming previous observations7–10,18. Eight of the nine Rv3806c mutations were also associated with increased ethambutol resistance (Supplementary Table 3), although this association was only statistically significant for the Rv3806c mutations affecting codons 237 (P = 0.014) and 240 (P = 0.042) and for all Rv3806c mutations taken as a group (P = 0.001) (Table 2), possibly because some of the individual Rv3806c mutations were relatively uncommon in our clinical strain sample. Rv3806c mutations were most frequently present in strains with high-level ethambutol resistance, present in 9 of 15 isolates (60%) with an ethambutol MIC of >16 µg/ml compared to in 2 of 28 isolates (7%) with an ethambutol MIC of 4–16 µg/ml and in 3of 20 isolates (15%) with an ethambutol MIC of <4 µg/ml (P < 0.001). In a model that included selected embB mutations and any Rv3806c mutation, Rv3806c mutations increased the ethambutol MIC by 1.4-fold over the MIC conferred by embB mutations alone (P = 0.009; 95% confidence interval (CI) of 1.1-fold to 1.7-fold) (Table 2). The upper boundary of this confidence interval is close to the twofold increase in MIC conferred by Rv3806c mutations upon allelic exchange (see below). None of the mutations in Rv3792 (synonymous or nonsynonymous), nuoD or embC were significantly associated with ethambutol resistance.
Table 2.
Genes, selected mutations and ethambutol MICs in clinical M. tuberculosis isolates
| Gene | Encoded alteration | Fold increase in ethambutol MIC (µg/ml)a (95% CI) |
P valueb |
|---|---|---|---|
| embB nonsynonymous mutations | |||
| All wild type/phylogeneticc | 1.4 (1.2–1.8) | ||
| embB codon 306 | p.Met306Ile | 7.8 (5.7–10.9) | <0.001 |
| p.Met306Val | 12.4 (9.5–16.3) | <0.001 | |
| embB codon 354 | p.Asp354Ala | 3.9 (1.9–8.1) | <0.001 |
| embB codon 406 | p.Gly406Asp | 5.0 (3.3–7.6) | <0.001 |
| p.Gly406Cys | 7.8 (3.8–16.2) | <0.001 | |
| p.Gly406Ser | 7.8 (3.8–16.2) | <0.001 | |
| embB codon 497 | p.Gln497Arg | 7.8 (5.7–10.9) | <0.001 |
| Rv3806c nonsynonymous mutations | |||
| All wild type/phylogenetic | 4.2 (3.0–5.8) | ||
| Rv3806c codon 237 | p.Arg237Cys | 5.4 (1.4–20.9) | 0.014 |
| Rv3806c codon 240 | p.Arg240Cys | 5.4 (1.1–27.8) | 0.042 |
| Rv3806c any nonsynonymous mutations | |||
| All wild type/phylogenetic | 4.1 (2.8–5.9) | ||
| Rv3806c | Any nonsynonymous change |
3.6 (1.6–8.1) | 0.001 |
| Rv3792 synonymous or nonsynonymous mutations | |||
| All wild type/phylogenetic | 5.3 (3.6–7.8) | ||
| Rv3792 | Any synonymous or nonsynonymous change |
0.7 (0.3–2.1) | 0.55 |
| embC nonsynonymous mutations | |||
| All wild type/phylogenetic | 5.0 (3.5–7.2) | ||
| embC codon 779 | p.Ala779Val | 2.3 (0.3–15.6) | 0.40 |
| nuoD nonsynonymous mutations | |||
| All wild type/phylogenetic | 5.3 (3.7–7.6) | ||
| nuoD codon 428 | p.Val428Ile | –d | >0.999 |
| Select embB mutations and any nonsynonymous Rv3806c mutation | |||
| All wild type/phylogenetic | 1.4 (1.2–1.8) | ||
| embB codon 306 | p.Met306Ile | 7.9 (5.8–10.6) | <0.001 |
| p.Met306Val | 11.1 (8.7–14.2) | <0.001 | |
| embB codon 354 | p.Asp354Ala | 3.9 (1.9–7.9) | <0.001 |
| embB codon 406 | p.Gly406Asp | 4.6 (3.1–6.6) | <0.001 |
| p.Gly406Cys | 7.9 (3.9–15.9) | <0.001 | |
| p.Gly406Ser | 5.8 (2.8–12.1) | <0.001 | |
| embB codon 497 | p.Gln497Arg | 7.4 (5.4–10.1) | <0.001 |
| embB codon 1082 | p.Thr1082Ala | 2.0 (1.2–3.3) | 0.011 |
| Rv3806c | Any nonsynonymous change |
1.4e (1.1–1.7) | 0.009 |
Fold increase in the geometric mean ethambutol MIC compared to the MIC for the group comprising wild-type strains and those with SNPs representing phylogenetic markers (wild type/phylogenetic). For example, the geometric mean ethambutol MIC for the EmbB p.Met306Ile alteration is 1.4 × 7.8 = 10.92 µg/ml, whereas, for the Rv3806c p.Arg237Cys alteration, the geometric mean ethambutol MIC is 4.2 × 5.4 = 22.68 µg/ml.
Pairwise P values unadjusted for multiple comparisons comparing strains with mutations to the wild-type/phylogenetic strains.
Reverse-transformed intercept. This provides the MIC for those without mutations at any of the codons included in the model; for example, in the first model, the estimated mean MIC for an individual without embB mutations at codons 306, 354, 406, 497, 853, 1073 and 1082 is 1.4 µg/ml.
When CIs were broad, parameter estimates were omitted but P values are provided.
Additional fold increase in ethambutol MIC in a model with select embB and Rv3806c mutations. For example, the geometric mean MIC for an M. tuberculosis isolate containing both an EmbB p.Met306Val alteration and an Rv3806c alteration (any nonsynonymous change) would be 1.4 × 11.1 × 1.4 = 21.76 µg/ml.
Transfer of Rv3806c, Rv3792, embC and nuoD mutations into a drug-susceptible strain and into its isogenic embB-mutant strain
We introduced different combinations of nuoD, Rv3792 and Rv3806c mutations identified in clinical and laboratory ethambutol-resistant strains into the genomes of the susceptible M. tuberculosis 210 strain and the ethambutol-resistant NJT210GTG strain that encodes a mutation in codon 306 of embB to determine the effect of each mutation on ethambutol resistance (Table 3). Similar allelic exchange studies attempted with embC mutations were unsuccessful, despite multiple attempts. The amino acid change encoded in nuoD did not affect the ethambutol MICs of the wild-type and NJT210GTG strains. The introduction of Rv3806c mutations into either codon 188, 237, 240 or 249 caused the ethambutol MIC to double from 2 to 4 µg/ml in strain 210 and to increase from 16 to 32 µg/ml in strain NJT210GTG (Table 3). The latter ethambutol MIC would be classified by most groups as indicating high-level ethambutol resistance. We also reintroduced the wild-type Rv3806c sequence into two of the isogenic Rv3806c mutants, converting them back to the sequence of the parental 210 strain. This reintroduction of Rv3806c caused the ethambutol MIC to revert back to 2 µg/ml (Table 3). Thus, Rv3806c mutations cause wild-type ethambutol-susceptible strains to acquire elevated ethambutol MICs within the susceptible range (breakpoint of 5.0–7.5 µg/ml) and cause strains with a mutation in codon 306 of embB to increase their ethambutol MICs into the range representing high-level resistance (MIC > 16 µg/ml). Interestingly, the introduction of the synonymous mutation in Rv3792 encoding p.Leu198Leu increased the ethambutol MICs of the wild-type and Rv3806c-mutant strains by four- and twofold, respectively (Table 3). Furthermore, reintroducing the wild-type nucleotide into Rv3792 isogenic mutants caused ethambutol MICs to revert back to the original concentration of 2 µg/ml (Table 3). In contrast, introducing the same Rv3792 mutation into NJT210GTG did not change the ethambutol MIC. Thus, it seems that Rv3792 mutations can increase ethambutol MIC to 8 µg/ml in several strains with MICs of <8 µg/ml but these same mutations do not have any further affect when introduced into strains that already have MICs of ≥16 µg/ml. These results represent the first description to our knowledge of an association between a synonymous coding mutation and drug resistance in M. tuberculosis.
Table 3.
Ethambutol MICs of isogenic and overexpression strains
| Strain | Inserted mutation | Ethambutol MIC (µg/ml)c |
||||
|---|---|---|---|---|---|---|
| embBa | Rv3806ca | Rv3792a | nuoDa | pMV261-Rv3806cb | ||
| 210 | WT | WT | WT | WT | NA | 2 |
| 210::Rv3806c-188 | WT | p.Val188Ala | WT | WT | NA | 4 |
| 210::Rv3806c-237 | WT | p.Ala237Val | WT | WT | NA | 4 |
| 210::Rv3806c-240 | WT | p.Arg240Cys | WT | WT | NA | 4 |
| 210::Rv3806c-249 | WT | p.Ala249Gly | WT | WT | NA | 4 |
| 210::Rv3792-198 | WT | WT | p.Leu198Leu | WT | NA | 8 |
| 210::Rv3806c-237/Rv3792-198 | WT | p.Ala237Val | p.Leu198Leu | WT | NA | 8 |
| 210::nuoD-287 | WT | WT | WT | p.Ala287Ser | NA | 2 |
| 210::Rv3806c-237-WT | WT | WT | WT | WT | NA | 2 |
| 210::Rv3806c-249-WT | WT | WT | WT | WT | NA | 2 |
| 210::Rv3792-198-WT | WT | WT | WT | WT | NA | 2 |
| 210::Rv3806c-237/Rv3792-198-WT | WT | p.Ala237Val | WT | WT | NA | 4 |
| NJT210GTG | p.Met306Val | WT | WT | WT | NA | 16 |
| NJT210GTG::Rv3806c-237 | p.Met306Val | p.Ala237Val | WT | WT | NA | 32 |
| NJT210GTG::Rv3806c-240 | p.Met306Val | p.Arg240Cys p.Gly269Gly | WT | WT | NA | 32 |
| NJT210GTG::Rv3806c-249 | p.Met306Val | p.Ala249Gly | WT | WT | NA | 32 |
| NJT210GTG::Rv3792-198 | p.Met306Val | WT | p.Leu198Leu | WT | NA | 16 |
| NJT210GTG::Rv3806c-237/Rv3792-198 | p.Met306Val | p.Ala237Val | p.Leu198Leu | WT | NA | 32 |
| NJT210GTG::nuoD-287 | p.Met306Val | WT | WT | p.Ala287Ser | NA | 16 |
| NJT210GTG::embB-306-WT | WT | WT | WT | WT | NA | 2 |
| 210::pMV261 | WT | WT | WT | WT | NA | 2 |
| 210::pMV261-Rv3806c-WT | WT | WT | WT | WT | WT | 4 |
| 210::pMV261-Rv3806c-188 | WT | WT | WT | WT | p.Val188Ala | 4 |
| 210::pMV261-Rv3806c-237 | WT | WT | WT | WT | p.Ala237Val | 4 |
| 210::pMV261-Rv3806c-240 | WT | WT | WT | WT | p.Arg240Cys | 4 |
| 210::pMV261-Rv3806c-249 | WT | WT | WT | WT | p.Ala249Gly | 4 |
| NJT210GTG::pMV261 | p.Met306Val | WT | WT | WT | NA | 16 |
| NJT210GTG::pMV261-Rv3806c-WT | p.Met306Val | WT | WT | WT | WT | 32 |
| NJT210GTG::pMV261-Rv3806c-188 | p.Met306Val | WT | WT | WT | p.Val188Ala | 32 |
| NJT210GTG::pMV261-Rv3806c-237 | p.Met306Val | WT | WT | WT | p.Ala237Val | 32 |
| NJT210GTG::pMV261-Rv3806c-240 | p.Met306Val | WT | WT | WT | p.Arg240Cys | 32 |
| NJT210GTG::pMV261-Rv3806c-249 | p.Met306Val | WT | WT | WT | p.Ala249Gly | 32 |
WT, wild type, no mutation inserted; NA, not applicable.
Protein alterations encoded by point mutations inserted in the chromosomal genes.
Overexpression of different Rv3806c alleles.
Ethambutol MIC determined by BACTEC 460TB and 7H10 agar proportion. MIC measurements were performed in triplicate by each method; all six replicates produced identical values (as shown) for each strain. MICs for isoniazid (0.05 µg/ml) and/or rifampin (0.05 µg/ml) were identical for all strains listed.
Effects on replicative fitness
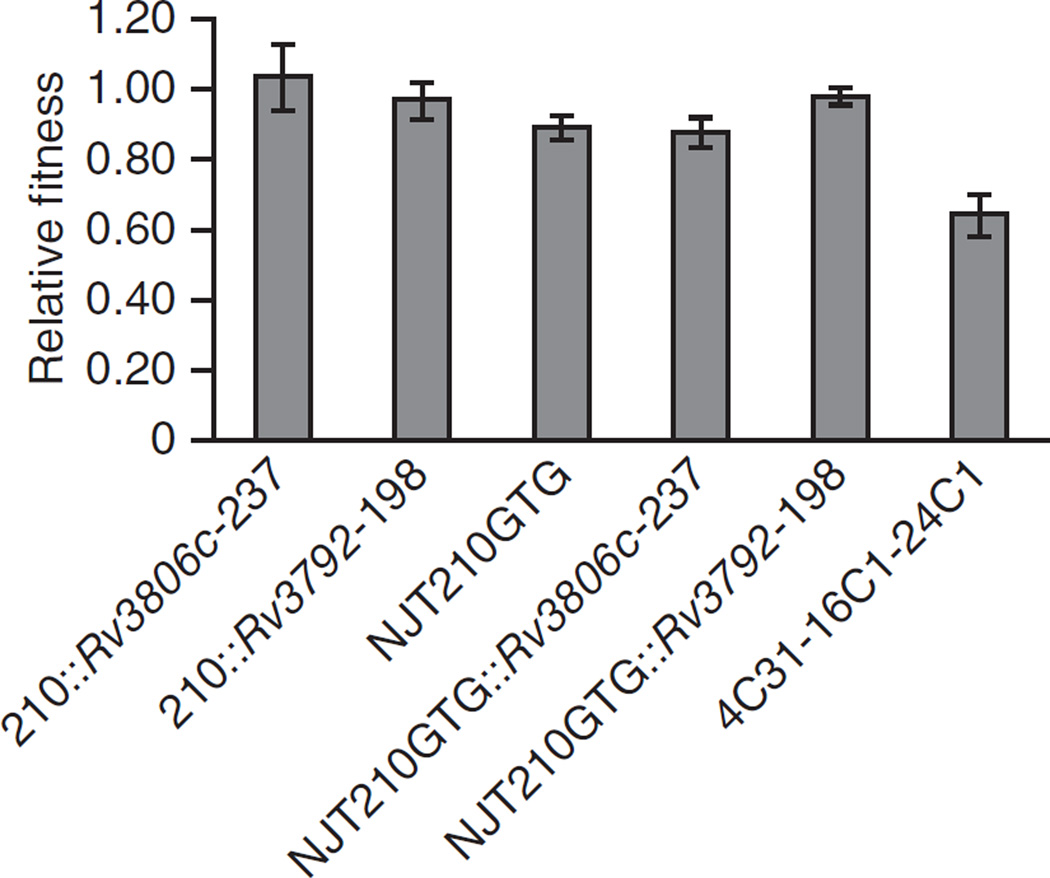
Recent studies have demonstrated that mutations in rpoB conferring rifampin resistance can affect strain fitness in M. tuberculosis and that compensatory mutations can develop19. We examined the fitness costs of embB, Rv3806c and Rv3792 mutations in pairwise competition experiments20. The isogenic Rv3806c- and Rv3792-mutant strains had no decrease in fitness compared to the parental 210 strain (Fig. 3). However, a fitness cost was observed for the ethambutol-resistant strain with a mutation in codon 306 of embB. The fitness cost of the embB codon 306 mutation could be compensated by the addition of the Rv3792 synonymous mutation but not by the addition of the Rv3806c mutation. However, it is noteworthy that the addition of the Rv3806c mutation to the background of embB codon 306 mutation did not reduce fitness further. All of the isogenic strains showed similar growth curves in 7H9 medium compared to the 210 strain (Supplementary Fig. 2a). These results suggest that Rv3806c mutations do not occur in conjunction with embB codon 306 mutations in clinical M. tuberculosis to compensate for decreased fitness. Rather, Rv3806c mutations serve to further increase the ethambutol MIC, which is accomplished without the mutations imposing additional fitness costs on the M. tuberculosis bacillus.
Figure 3.
Relative competitive reductions in fitness for isogenic embB, Rv3806c and/or Rv3792 mutants and strain 4C31-16C1-24C1. Each test strain was mixed at approximately a 1:1 ratio with the parental 210 strain and then cultured in the absence of ethambutol to stationary phase. Confirmed initial (inoculation) and final CFUs were used to calculate the relative fitness of each strain compared to strain 210. The means and s.d. of three independent experiments are shown. The results show a fitness cost for the embB306 mutation but not for the other mutations introduced by allelic exchange. The multiple-step mutant strain 4C31-16C1-24C1 also has reduced fitness.
Ethambutol does not directly inhibit Rv3806c
An obvious explanation for the ethambutol resistance observed in Rv3806c-mutant strains is that the Rv3806c protein is a target for ethambutol, in a manner similar to that described for the Emb proteins. If true, overexpression of wild-type Rv3806c should increase ethambutol MIC moderately, and overexpression of mutant Rv3806c should result in a dramatic increase in ethambutol MIC5. We overexpressed wild-type Rv3806c as well as Rv3806c codon 188, 237, 240 and 249 mutants in strains 210 and NJT210GTG (with expression confirmed by quantitative RT-PCR (qRT-PCR); Supplementary Fig. 3). Overexpression of Rv3806c doubled parental ethambutol MICs, regardless of whether the wild-type or mutant Rv3806c sequence was used (Table 3).
To directly assess whether ethambutol inhibits the enzymatic activity of the Rv3806c protein, we tested membrane extracts from an Escherichia coli strain that overexpresses recombinant M. tuberculosis Rv3806c15 for phosphoribosyl transferase activity in the presence of various concentrations of ethambutol. We did not detect any inhibition of DPPR synthase activity, even at the highest ethambutol concentration of 50 µg/ml, demonstrating that ethambutol does not alter Rv3806c activity (Supplementary Fig. 4a,b). In contrast, in vitro analysis of arabinosyl transferase activities clearly showed inhibition in the presence of ethambutol21. Combined with the overexpression data, these results suggest that Rv3806c is unlikely to be a target of ethambutol.
Rv3806c mutations increase DPA levels
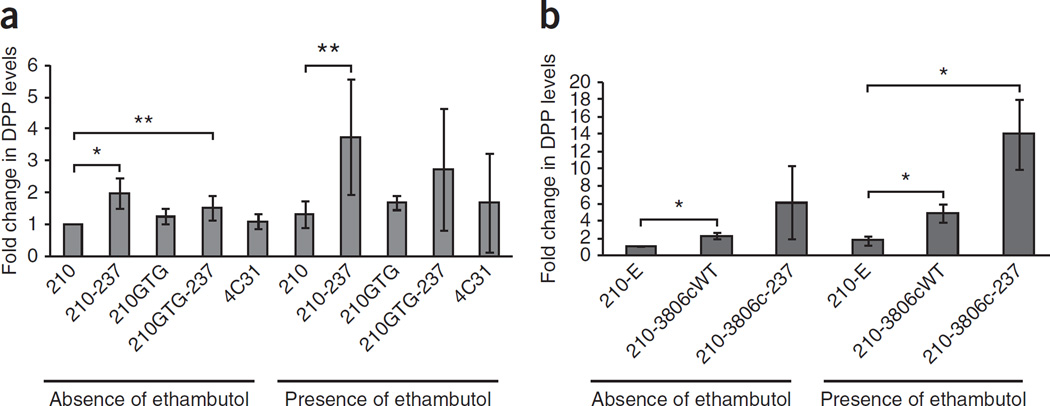
We investigated the possibility that Rv3806c mutations cause ethambutol resistance by increasing DPA levels rather than by altering the ability of ethambutol to inhibit Rv3806c. Our hypothesis was that increased DPA levels would competitively inhibit the binding of ethambutol to its Emb target, thereby increasing ethambutol MICs. The high-performance liquid chromatography (HPLC) and mass spectrometry method used could not distinguish decaprenylmonophosphoryl-β-d-ribose (DPR) from DPA, as they coeluted on the HPLC column; therefore, the combined levels of both DPR and DPA (DPP) were measured instead. We measured DPP levels in cellular extracts from strains 210 and NJT210GTG, these two strains with a mutation in Rv3806c encoding a p.Ala237Val substitution and the 210 strain overexpressing wild-type or mutant Rv3806c. Strain 4C31 was used as an additional control because its MIC is 4 µg/ml although its Rv3806c gene is wild type. Both strains with wild-type Rv3806c sequences (4C31 and NJT210GTG) showed no changes in DPP levels compared to strain 210 (Fig. 4a). In contrast, DPP levels were significantly higher in the strains with the Rv3806c mutation encoding p.Ala237Val, both in the wild-type and embB codon 306 mutant backgrounds. Ethambutol treatment further increased DPP levels in the Rv3806c mutants compared to the ethambutol-treated 210 strain, although interexperimental variability prevented the increase from reaching statistical significance in the ethambutol-treated Rv3806c-mutant strain that also contained an embB codon 306 mutation (P = 0.08) (Fig. 4a). If Rv3806c mutations result in an increase in function that elevates DPP levels, then overexpression of wild-type Rv3806c should also increase DPP levels. We found that this was indeed the case (Fig. 4b), a result that also is consistent with the increased ethambutol MIC conferred by overexpressing this gene. DPP levels were further increased in the strain with overexpression of mutant Rv3806c and upon treatment with ethambutol.
Figure 4.
Role of Rv3806c in ethambutol resistance. (a) DPP (DPA + DPR) levels in strains with mutant Rv3806c, embB and Rv3792. Fold increases in normalized nanograms of DPP per gram of bacterial cells are shown in various strains grown in the presence and absence of ethambutol. A significant increase in DPP levels is observed in the Rv3806c-mutant strains except for in 210GTG-237 treated with ethambutol. (b) Effect of Rv3806c overexpression on DPP levels. For a,b, the strains used included the following: 210, wild-type strain 210; 210–237, strain 210::Rv3806c-237 (strain 210 containing the Rv3806c p.Ala237Val alteration); 210GTG, NJT210GTG (strain 210 containing the EmbB p.Met306Val alteration); 210GTG-237, NJT210GTG::Rv3806c-237 (strain 210 containing both EmbB p.Met306Val and Rv3806c p.Ala237Val alterations); 210-E, strain 210::pMV261 (strain 210 containing the plasmid pMV261); 210-Rv3806cWT, strain 210::pMV261-Rv3806c-WT (strain 210 overexpressing wild-type Rv3806c); 210-Rv3806c-237, strain 210::pMV261-Rv3806c-237 (strain 210 overexpressing mutant Rv3806c Ala237Val). Significant differences in DPP levels were calculated by Student’s t test: *P = 0.01, **P = 0.04. Means and s.d. of three independent experiments are shown.
Synonymous mutations in Rv3792 increase embC expression
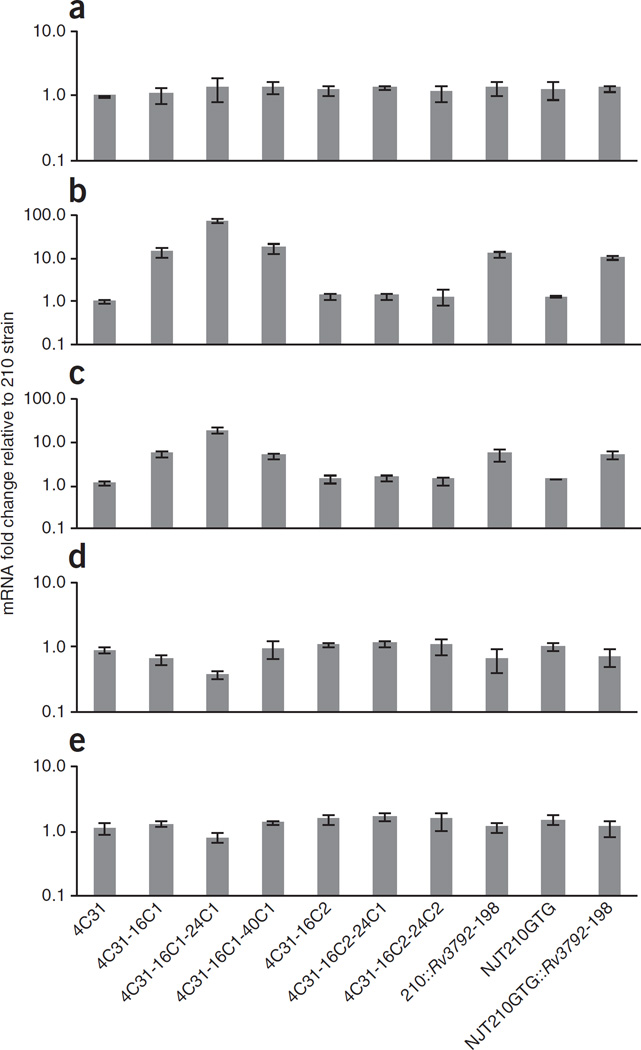
EmbC is a known ethambutol target5,22. Furthermore, overexpression of Mycobacterium smegmatis embC in M. tuberculosis increases the ethambutol MIC, although overexpression of M. tuberculosis embC in M. tuberculosis has not been achieved directly by our group (data not shown) or others, possibly because of toxic effects22. Noting that Rv3792 is situated immediately upstream of embC, we studied whether the increase in ethambutol MIC observed in our strains with a synonymous mutation in Rv3792 was caused by increased embC expression. We performed qRT-PCR studies of Rv3792, embC, embB and Rv3806c in the laboratory-constructed isogenic Rv3792-mutant strains as well as in all the strains shown in Figure 2 (Fig. 5). For Rv3792, two different RT-PCR assays were performed: one measuring mRNA expression upstream of the Rv3792 mutation (5′ Rv3792) and the other measuring mRNA expression downstream of the mutation (3′ Rv3792). All strains had similar levels of 5′ Rv3792 expression (Fig. 5a). However, 3′ Rv3792 expression was 15-fold higher in the Rv3792-mutant strains, except for in the highly ethambutol-resistant 4C31-16C1-24C1 strain, which showed an approximately 80-fold increase in expression (Fig. 5b). Expression of embC was also elevated in the Rv3792 mutant strains by approximately 19-fold in 4C31-16C1-24C1 and by approximately 5-fold in the other mutant strains (Fig. 5c). All other genes studied were expressed at similar levels in all strains, except for embB, whose mRNA levels were decreased by 2.5-fold in strain 4C31-16C1-24C1 (P < 0.01) (Fig. 5d,e). Strain 4C31-16C1-24C1 showed a strong decrease in fitness and slow growth compared to other strains (Fig. 3 and Supplementary Fig. 2b), suggesting that very high Rv3792 downstream expression may be one factor contributing to this complex phenotype. Notably, Rv3806c expression was not increased in any of the Rv3806c-mutant strains, confirming that Rv3806c and Rv3792 mutant-strains have different underlying resistance mechanisms (Fig. 5e). We observed that the Rv3792 synonymous mutation was situated 10 nt upstream of a codon for methionine at amino acid position 202 (ref. 11). To examine the direct effect of increased Rv3792 expression on ethambutol resistance, we overexpressed both the entire Rv3792 ORF as well as a truncated version of Rv3792 encoding protein starting at Met202. Ethambutol MICs did not increase in either strain 210 or NJT210GTG expressing either construct, although overexpression was confirmed by qRT-PCR (data not shown). These results strongly suggest that synonymous Rv3792 mutations cause ethambutol resistance by increasing embC expression but not by increasing 3′ Rv3792 expression, probably through activating an internal promoter within the Rv3792 gene.
Figure 5.
Effect of Rv3792 synonymous mutations on Rv3792 and embC mRNA levels. RNA was extracted from cultures grown to mid-log phase. Transcript levels were analyzed by qRT-PCR, and fold changes were determined by comparison to levels in the 210 strain after normalization to 16S rRNA levels. (a–e) Relative mRNA levels of 5′ Rv3792 (a), 3′ Rv3792 (b), embC (c), embB (d) and Rv3806c (e). Means and s.d. are derived from three independent experiments. Large increases in the expression of 3′ Rv3792 and of embC are seen only in the Rv3792 mutants. No strain has increased Rv3806c expression; embB expression is mildly decreased in strain 4C31-16C1-24C1.
DISCUSSION
This study describes the first known cause of high-level ethambutol resistance. It also describes to our knowledge the first association between a gain-of-function mutation (in the case of Rv3806c) or a synonymous SNP (in the case of Rv3792) and drug resistance in M. tuberculosis. Our findings suggest that the pathway to ethambutol resistance can be complex, involving mutations in several genes that interact to determine the eventual ethambutol MIC (Supplementary Fig. 5). All of the confirmed mutations conferring ethambutol resistance discovered in our study seemed to affect DPA synthesis or DPA utilization pathways. However, our clinical data and the mutations noted in strains 8C1 and 8C6, which did not increase ethambutol MICs in liquid medium but did modestly increase MICs on solid medium, suggest that other mechanisms of ethambutol resistance remain to be discovered. We suggest that canonical mutations in embB are common first-step mutations in the development of ethambutol resistance. However, mutations in non-canonical embB loci and perhaps in Rv3806c, embC, Rv3792 and other unknown genes may also occur as the first step in the evolution of ethambutol resistance. In these cases, the first mutation may not seem to increase the ethambutol MIC above the breakpoint of 5.0–7.5 µg/ml that defines clinical ethambutol resistance. These M. tuberculosis mutant strains will not be distinguished from wild-type isolates in clinical microbiology laboratories that perform standard phenotypic testing for drug susceptibility. Yet, these mutations may be important, as they represent the first, often pre-resistant step on the path to high-level ethambutol resistance.
Rv3806c is an essential gene for mycobacterial growth and viability15. Our study provides both indirect and direct evidence that ethambutol does not inhibit Rv3806c enzymatic activity, strongly suggesting that it is not itself a drug target for ethambutol. Instead, Rv3806c mutations seem to cause ethambutol resistance by increasing intracellular DPA levels. DPA may compete with ethambutol for Emb binding sites, and increased DPA levels in Rv3806c-mutant strains may preserve Emb enzymatic function in the presence of ethambutol. Simultaneous overproduction of DPA (by Rv3806c mutation) and inhibition of its use (by ethambutol treatment23,24) would be expected to result in the highest levels of DPA, as confirmed in our study. Rv3806c mutations likely elevate DPA levels by increasing the rate of decaprenyl phosphate conversion into DPPR, a precursor of DPA (Supplementary Fig. 1). However, the mechanism by which Rv3806c mutations lead to this increased conversion is unclear, and other mechanisms are also possible.
We did not find any Rv3792 mutations associated with ethambutol resistance in clinical M. tuberculosis strains, despite the fact that the synonymous mutation increased the ethambutol MIC in laboratory strains. The SNP in Rv3792 seemed to confer resistance by increasing transcription of embC, as overexpression of either the entire Rv3792 gene or its downstream portion did not affect the ethambutol MIC. We postulate that the Rv3792 SNP increased gene expression by activating an internal promoter. This mechanism is different from those previously described for synonymous mutations with physiological effects, which include biases in codon usage25,26 or coding sequence determinants27. Our results also suggest that mutations in embC can combine with mutations in embB and Rv3806c to produce strains with very high ethambutol MICs. We did not find any clinical isolates with high-level resistance to ethambutol that had embC mutations, consistent with published studies showing that embC mutations are found rarely in clinical strains and are mostly associated with embB mutations10. However, embC-mutant ethambutol-resistant strains have previously been identified in other clinical studies, and good evidence exists to suggest that both EmbB and EmbC are inhibited by ethambutol5,9,22. Although we did not find any mutations in addition to those in embB and Rv3792 in strain 4C31-16C1-24C1 that might explain its high-level ethambutol resistance, it is likely that the high level of embC expression that we observed in this strain is at least one contributing factor to its high-level resistance and unique phenotype. The transcriptional regulation of the embCAB operon is incompletely understood5,28,29. Experiments using our isogenic Rv3792 mutants are in progress to study the mechanism of embC regulation and to characterize the new Rv3792 internal promoter and its relationship with high-level resistance.
Our study contradicts commonly held beliefs that drug resistance in M. tuberculosis is caused by single-step mutations. These conclusions are clinically relevant, especially if they are found to be applicable to other tuberculosis drugs. In fact, several studies have identified clinical M. tuberculosis strains that carry the same resistance-conferring mutations but nonetheless exhibit varying MICs for rifampin or isoniazid30–32. We suggest that mutant strains with low-level resistance, such as the ones detected in our study, may be preferentially selected in patients exposed to drug concentrations below the MIC. As these mutant strains accumulate, they would constitute a pool from which fully drug-resistant strains could preferentially emerge. Our study also demonstrates the potential inadequacies of current methods for testing drug resistance in M. tuberculosis. Standard phenotypic testing uses breakpoint concentrations to define drug resistance. This approach cannot identify strains with low-level increases in MIC. The mutations targeted in molecular assays for drug resistance were originally identified by phenotypic testing for drug resistance. Thus, the molecular assays currently available would also fail to detect mutant strains with low-level resistance. Our study suggests that molecular tests for drug resistance should be broadened to include screening for mutations that can confer low-level resistance as well as for mutations that can cause high-level resistance. This strategy could provide important actionable information in the clinic to increase a drug dose, include an additional drug or switch to alternate therapy. Further investigations are required to determine whether our discoveries are more broadly applicable to other types of drug resistance.
ONLINE METHODS
Bacterial strains, culture conditions and susceptibility testing
M. tuberculosis strains were cultured as described previously33. MICs were determined both by 7H10 agar proportion and by radiometric BACTEC 460TB methods (Becton Dickinson), following the manufacturer’s instructions with minor modifications7. MICs determined by both methods showed a correlation of 92%; thus, unless stated otherwise in the text, MICs corresponded to those determined using BACTEC 460TB methods. All MICs were determined in triplicate. Because each value within a triplicate MIC test was almost always identical to the other values within the same triplicate set, a single MIC value is shown without standard deviation for each test unless otherwise indicated. For all plasmid construction, E. coli strains Top10 (Invitrogen) were grown in Luria-Bertani medium supplemented with 50 µg/ml kanamycin, where appropriate. Clinical M. tuberculosis strains with varying ethambutol MICs were selected from a pedigreed collection of 229 highly characterized M. tuberculosis isolates established by the World Health Organization Special Programme for Research and Training in Tropical Disease using geographic, phylogenetic and ethambutol MIC diversity as selection criteria17.
DNA isolation, PCR, qRT-PCR and bidirectional DNA sequencing
Isolation of genomic DNA and PCR were performed as described previously33,34. Rv3806c, Rv3792, embC and embB mRNA expression levels were measured by qRT-PCR, and ratios of their mRNA levels to corresponding levels in the 210 strain were determined after normalization to 16S rRNA levels, as described previously33,35. Direct Sanger sequencing of PCR products was performed with a Dye Terminator kit on an ABI 3100 Genetic Analyzer (Applied Biosystems). Sequences for the primers and molecular beacons used in this study are available upon request.
Selection and isolation of ethambutol-resistant mutants
Single-step ethambutol-resistant mutants were isolated by plating 1 × 109 CFUs of ethambutol-susceptible M. tuberculosis strain 210 onto 7H10 medium containing 8, 16 or 32 µg/ml ethambutol, as described previously8. For multistep selection for ethambutol resistance, first-step ethambutol-resistant mutants were isolated in the same manner as single-step mutants, except that plating was carried out on medium with 4 µg/ml ethambutol. Second-step mutant strains were created by isolating individual colonies of first-step mutant strains, culturing in 7H9 medium without ethambutol and then plating 1 × 109 CFUs on medium with 16 µg/ml ethambutol. This process was repeated to generate third-step mutants, except that this time the culture was plated on medium with either 24, 32, 40, 48 or 56 µg/ml ethambutol.
Whole-genome sequencing and mutation detection of in vitro–selected strains
Whole-genome sequences were generated on a 454 GS FLX system (Roche) using Titanium according to the manufacturer’s instructions (Roche). The sequencing platform covered approximately 91 to 98% of the M. tuberculosis genome with an average coverage ranging from 12× to 19×, similar to the coverage reported in recent studies36. Base calling was performed using the bundled 454 software. Initially, the GS De novo Assembler (Roche) was used to assemble the sequence reads from each strain. The best assembly was chosen on the basis of total sequence coverage, total contig length and number of contigs. The sequence reads from each isolate, including the reference isolate (210 strain), were assembled again using the GS Reference Mapper application (Roche). We identified polymorphisms by accepting all conflicts reported in the 454HCDiffs.txt file where at least 80% of the sequence reads at the affected location were in conflict with the reference assembly and in agreement with at least three non-duplicate reads. SNP accuracy was confirmed by 3′ mismatch qPCR. Final SNPs were confirmed by direct Sanger sequencing.
Isogenic strain construction and overexpression studies
Rv3806c, Rv3792 and nuoD sequences containing point mutations were substituted for wild-type sequences in M. tuberculosis strain 210 and/or M. tuberculosis strain NJT210GTG using allelic exchange techniques as described previously8. Mutated and wild-type Rv3806c and native and truncated Rv3792 were also overexpressed from pMV261 under the control of the hsp60 promoter35.
Competitive fitness assays
Fitness costs were determined by pairwise competition assays in the absence of ethambutol, performed as previously described with minor modifications20. Strains were mixed and inoculated at approximately 4 × 104 total CFUs, and mixtures were grown to stationary phase. Mixed cultures were then serially diluted and plated in 7H10 medium. Colonies from the baseline and end-point cultures were determined to be wild type or mutant using appropriate molecular beacon assays. Finally, the fitness cost was calculated as described previously20.
Phosphoribose transferase assays
To test the effect of ethambutol on Rv3806c (DPPR synthase), the M. tuberculosis Rv3806c protein was expressed and purified from E. coli strain ER2556 (New England Biolabs) as described previously15. A standard 30-µl reaction contained 45 nCi purified substrate phosphor[14C]ribosyl diphosphate (p[14C]Rpp), 4 µg of decaprenyl phosphate (Indofine Chemicals), 5 µg of membrane protein and 0.25% CHAPS (Sigma) in buffer A (50 mM MOPS, 1 mM magnesium chloride and 5 mM β-mercaptoethanol). Radiolabeled substrate p[14C]Rpp was prepared enzymatically from uniformly labeled [14C]glucose (American Radiolabeled Chemicals) as described previously37. The transferase assay reaction was incubated at 37 °C for 30 min and then stopped by the addition of chloroform and methanol. After centrifugation to generate two phases, an aliquot of the lower, organic phase was dried, and was radioactivity measured using a scintillation counter (Beckman Coulter, LS6500). To confirm the identity of the product, the remaining organic phase was dried under a slow stream of air and resuspended in chloroform and methanol. Samples were then spotted on a silica gel 60 TLC plate (Merck), and the plate was run in chloroform:methanol:concentrated ammonium hydroxide:1 M ammonium acetate:water and visualized using a phosphorimager (Molecular Dynamics). To determine the effect of ethambutol on the DPPR synthase, various concentrations of ethambutol were added to the standard reaction mixture.
Enzymatic conversion to DPA in different M. tuberculosis strains in the presence and absence of ethambutol
We measured the amount of arabinose in cellular DPA in the presence and absence of ethambutol using the wild-type 210 strain and various isogenic strains. These arabinoses levels served to indirectly measure DPA accumulation. Each strain was cultured to equivalent optical density. Equal volumes of each culture were then centrifuged, and pellets were resuspended in 20 volumes of chloroform and methanol, incubated for 3 h at room temperature and centrifuged again. The organic phase was evaporated to about 500 µl in volume, and an aliquot (100 µl) was transferred to a screw-cap gas chromatography vial to which 5 µg of the internal standard 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine-N-nonadecanoyl (Avanti Polar Lipids, Al) was added. An aliquot of each sample (1 µl) was then analyzed by liquid chromatography coupled with mass spectrometry. Under the liquid chromatography conditions used, DPA and DPR did not reliably separate, and DPP levels were thus calculated. The liquid chromatography and mass spectrometry (ESI/APCI ionization) analysis for DPP was carried out on an Agilent 6220 time-of-flight mass spectrometer equipped with a MultiMode Source (negative mode) and an Agilent 1200 binary pump. A modified liquid chromatography and mass spectrometry procedure from Shui et al. was followed38. The column used was a Waters XBridge (C18, 2.1 mm × 150 mm) with a particle size of 5 µM, and the column temperature was maintained at 45 °C. Separation was carried out using a gradient of solvent A (99% methanol and 1% 500 mM ammonium acetate) and solvent B (79% n-propanol, 20% hexane and 1% 500 mM ammonium acetate) from 100% solvent A to 100% solvent B. The drying gas temperature was 300 °C, and the vaporizer temperature was set at 200 °C. Typically, 5 µl of sample was injected for analysis, with a flow rate of 0.32 ml/min, and the total run time was 45 min. The fragmentor voltage was set to 120 V. The mass spectrum was acquired from m/z values of 250 to 3,200 Da with a frequency of one scan per second. Selected ions at m/z values of 922.63 Da (DPP) and 1,022.82 Da (internal standard) were integrated, and the ratio was computed. To convert to nanograms, a standard containing 100 ng of synthetic DPA and the same amount of internal standard was analyzed in the same fashion to yield a response factor. All DPP levels were first normalized by their wet weight because ethambutol substantially inhibited biomass formation in the treated versus the untreated cells. All experiments were performed at least three times using cultures of separate strains. To facilitate analysis across experiments, these results were then normalized to DPP levels in the wild-type 210 strain without ethambutol treatment in each experiment.
Statistical analysis
For the analysis of clinical isolates, testing for MIC differences by mutations was performed using regression models. Ethambutol MICs were assumed to be interval censored between the highest and lowest concentrations inhibiting growth and were log-transformed, assuming normal errors. Estimates and 95% CIs at each codon compare mutations to wild type or phylogenetic mutations. Estimates were reverse transformed, and all estimates are therefore relative to those for the group including wild-type strains and those with only phylogenetic mutations. For example, an estimate of 4 indicates that a mutation has an ethambutol MIC that is 4 times that of the group including wild-type strains and those with only phylogenetic mutations. Two-sided Wald P-value testing for any difference in MIC by mutation at a codon and pairwise comparisons of MIC values for specific mutations to those in the group including wild-type strains and those with only phylogenetic mutations are provided, unadjusted for multiple comparisons. The LIFEREG procedure in SAS 9.2 was used for fitting models. In the analysis of DPP levels in laboratory-generated mutants, fold increases in DPP levels were compared using a Student’s t test, as appropriate.
Acknowledgments
We thank S. Kuppasani for performing the direct Sanger sequencing. This work was supported in part by National Institute of Allergy and Infectious Diseases, US National Institutes of Health grants AI080653, AI065663 and AI037139 and by Pathogen Functional Genomics Resource Center contract N01-AI5447.
Footnotes
Accession codes. Whole-genome sequencing data for the strains 210, NJT210GTG, 8C1, 8C6, 16C10, 32C4, 4C31, 4C31-16C1, 4C31-16C2, 4C31-16C1-24C1, 4C31-16C1-40C1, 4C31-16C2-24C1 and 4C31-16C2-24C2 have been deposited in the NCBI Sequence Read Archive (SRA) under accessions SRX018836, SRS041973, SRS041969, SRS041970, SRS041975, SRS041974, SRS041977, SRS041978, SRS041979, SRS042019, SRS041999, SRS041980 and SRS041997, respectively.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
H.S., S.L., A.A., M.J., M.M., S.N.P., D.C., R.F. and D.A. designed experiments. H.S., S.L., A.A. and M.J. performed experiments. H.S., A.A., M.H., M.M., S.N.P., D.C., R.F. and D.A. performed analysis of results. S.K., D.A. and H.S. performed statistical analysis. H.S., S.K., M.M., D.C., R.F. and D.A. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Zignol M, et al. Global incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 2006;194:479–485. doi: 10.1086/505877. [DOI] [PubMed] [Google Scholar]
- 2.Sharma SK, Mohan A. Multidrug-resistant tuberculosis: a menace that threatens to destabilize tuberculosis control. Chest. 2006;130:261–272. doi: 10.1378/chest.130.1.261. [DOI] [PubMed] [Google Scholar]
- 3.Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 4.Riska PF, Jacobs WR, Alland D. Molecular determinants of drug resistance in tuberculosis. Int. J. Tuberc. Lung Dis. 2000;4:S4–S10. [PubMed] [Google Scholar]
- 5.Telenti A, et al. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 1997;3:567–570. doi: 10.1038/nm0597-567. [DOI] [PubMed] [Google Scholar]
- 6.Takiff HE, et al. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safi H, Sayers B, Hazbon MH, Alland D. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob. Agents Chemother. 2008;52:2027–2034. doi: 10.1128/AAC.01486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safi H, et al. Allelic exchange and mutant selection demonstrate that common clinical embCAB gene mutations only modestly increase resistance to ethambutol in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2010;54:103–108. doi: 10.1128/AAC.01288-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaswamy SV, et al. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2000;44:326–336. doi: 10.1128/aac.44.2.326-336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plinke C, et al. embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J. Antimicrob. Chemother. 2010;65:1359–1367. doi: 10.1093/jac/dkq120. [DOI] [PubMed] [Google Scholar]
- 11.Alderwick LJ, Seidel M, Sahm H, Besra GS, Eggeling L. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 2006;281:15653–15661. doi: 10.1074/jbc.M600045200. [DOI] [PubMed] [Google Scholar]
- 12.Alderwick LJ, et al. The C-terminal domain of the arabinosyltransferase Mycobacterium tuberculosis EmbC is a lectin-like carbohydrate binding module. PLoS Pathog. 2011;7:e1001299. doi: 10.1371/journal.ppat.1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikusová K, et al. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J. Bacteriol. 2005;187:8020–8025. doi: 10.1128/JB.187.23.8020-8025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mokrousov I, Otten T, Vyshnevskiy B, Narvskaya O. Detection of embB306 mutations in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis from northwestern Russia: implications for genotypic resistance testing. J. Clin. Microbiol. 2002;40:3810–3813. doi: 10.1128/JCM.40.10.3810-3813.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H, et al. Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-α-D-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-D-arabinose synthesis. J. Biol. Chem. 2005;280:24539–24543. doi: 10.1074/jbc.M504068200. [DOI] [PubMed] [Google Scholar]
- 16.Huang H, et al. Identification of amino acids and domains required for catalytic activity of DPPR synthase, a cell wall biosynthetic enzyme of Mycobacterium tuberculosis. Microbiology. 2008;154:736–743. doi: 10.1099/mic.0.2007/013532-0. [DOI] [PubMed] [Google Scholar]
- 17.Vincent V, et al. The TDR Tuberculosis Strain Bank: a resource for basic science, tool development and diagnostic services. Int. J. Tuberc. Lung Dis. 2012;16:24–31. doi: 10.5588/ijtld.11.0223. [DOI] [PubMed] [Google Scholar]
- 18.Starks AM, Gumusboga A, Plikaytis BB, Shinnick TM, Posey JE. Mutations at embB codon 306 are an important molecular indicator of ethambutol resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2009;53:1061–1066. doi: 10.1128/AAC.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comas I, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 2012;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagneux S, et al. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 21.Lee RE, Mikusova K, Brennan PJ, Besra GS. Synthesis of the arabinose donor. β-D-arabinofuranosyl-1-monophosphoryldecaprenol, development of a basic arabinosyl-transferase assay, and identification of ethambutol as an arabinosyl transferase inhibitor. J. Am. Chem. Soc. 1995;117:11829–11832. [Google Scholar]
- 22.Goude R, Amin AG, Chatterjee D, Parish T. The Arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2009;53:4138–4146. doi: 10.1128/AAC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolucka BA, McNeil MR, de Hoffmann E, Chojnacki T, Brennan PJ. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 1994;269:23328–23335. [PubMed] [Google Scholar]
- 24.Wolucka BA. Biosynthesis of d-arabinose in mycobacteria—a novel bacterial pathway with implications for antimycobacterial therapy. FEBS J. 2008;275:2691–2711. doi: 10.1111/j.1742-4658.2008.06395.x. [DOI] [PubMed] [Google Scholar]
- 25.dos Reis M, Wernisch L, Savva R. Unexpected correlations between gene expression and codon usage bias from microarray data for the whole Escherichia coli K-12 genome. Nucleic Acids Res. 2003;31:6976–6985. doi: 10.1093/nar/gkg897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgs PG, Ran W. Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage. Mol. Biol. Evol. 2008;25:2279–2291. doi: 10.1093/molbev/msn173. [DOI] [PubMed] [Google Scholar]
- 27.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goude R, Amin AG, Chatterjee D, Parish T. The critical role of embC in Mycobacterium tuberculosis. J. Bacteriol. 2008;190:4335–4341. doi: 10.1128/JB.01825-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escuyer VE, et al. The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J. Biol. Chem. 2001;276:48854–48862. doi: 10.1074/jbc.M102272200. [DOI] [PubMed] [Google Scholar]
- 30.Ohno H, Koga H, Kohno S, Tashiro T, Hara K. Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob. Agents Chemother. 1996;40:1053–1056. doi: 10.1128/aac.40.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang H-Y, et al. Characterization of rifampicin-resistant Mycobacterium tuberculosis in Taiwan. J. Med. Microbiol. 2003;52:239–245. doi: 10.1099/jmm.0.05045-0. [DOI] [PubMed] [Google Scholar]
- 32.Kim S-Y, et al. Molecular analysis of isoniazid resistance in Mycobacterium tuberculosis isolates recovered from South Korea. Diagn. Microbiol. Infect. Dis. 2003;47:497–502. doi: 10.1016/s0732-8893(03)00132-9. [DOI] [PubMed] [Google Scholar]
References
- 33.Safi H, et al. IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis. Mol. Microbiol. 2004;52:999–1012. doi: 10.1111/j.1365-2958.2004.04037.x. [DOI] [PubMed] [Google Scholar]
- 34.van Embden JD, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colangeli R, et al. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-kike protein Lsr2 in M. tuberculosis. PLoS Pathog. 2007;3:e87. doi: 10.1371/journal.ppat.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Supply P, et al. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat. Genet. 2013;45:172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherman MS, et al. Polyprenylphosphate-pentoses in Mycobacteria are synthesized from 5-phosphoribose pyrophosphate. J. Biol. Chem. 1996;271:29652–29658. doi: 10.1074/jbc.271.47.29652. [DOI] [PubMed] [Google Scholar]
- 38.Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. Sensitive profiling of chemically diverse bioactive lipids. J. Lipid Res. 2007;48:1976–1984. doi: 10.1194/jlr.M700060-JLR200. [DOI] [PubMed] [Google Scholar]