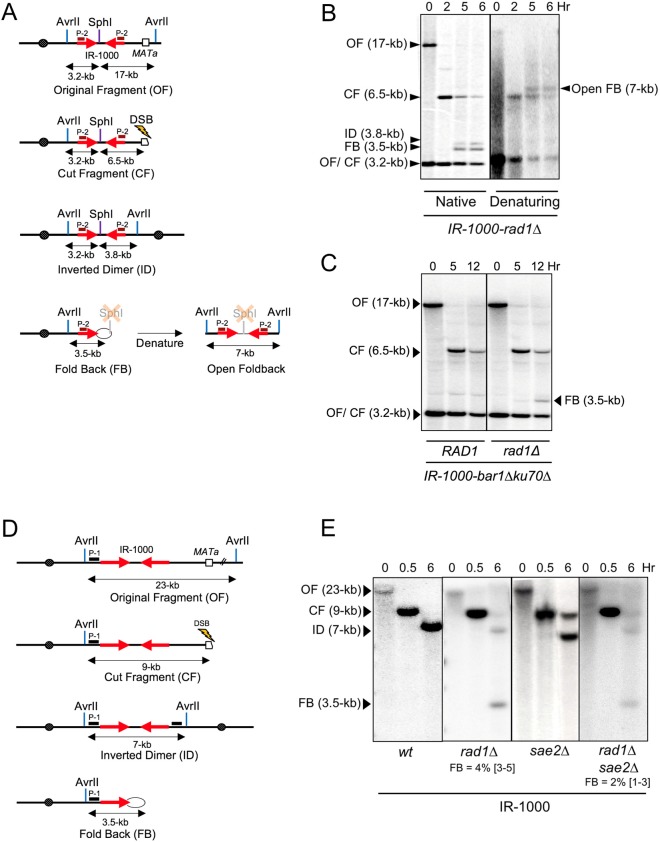
Fig 3. The role of Rad1 in processing of fold-backs with 1-kb spacers.
(A) The schematics of AvrII /SphI digest of chromosome III in IR-1000 (OF) and its derivatives, including: CF, ID, FB, and “open” fold-back resulting from denaturation of FB. The location of probe P-2, specific to PHO87 sequence, is indicated by brown box. (B) Southern blot analysis of DSB repair in rad1Δ derivative of IR-1000 strain following native or denaturing gel electrophoresis after AvrII /SphI digest of DNA isolated before (0 hr), or 2, 5, 6 hours after DSB induction, followed by hybridization with probe P-2. (C) FB formation in IR-1000 arrested at G1 stage of the cell cycle. DNA extracted from RAD1 or rad1Δ at 0-hr (prior to DSB induction) and 5 or 12-hr post-DSB-induction, digested with AvrII and SphI followed by native gel electrophoresis and hybridization with probe P-2. (D) Schematics of AvrII digested ChrIII and its derivatives; OF, CF, ID and FB in IR-1000. (E) Analysis of FB and ID formation in IR-1000 (wt) and its derivatives by AvrII digest, native gel electrophoresis, and hybridization with probe P-1. The median efficiencies of FB formation (%) and the range of the median [in brackets] are indicated (see S1 Fig and Materials and Methods for details).