Figure 1.
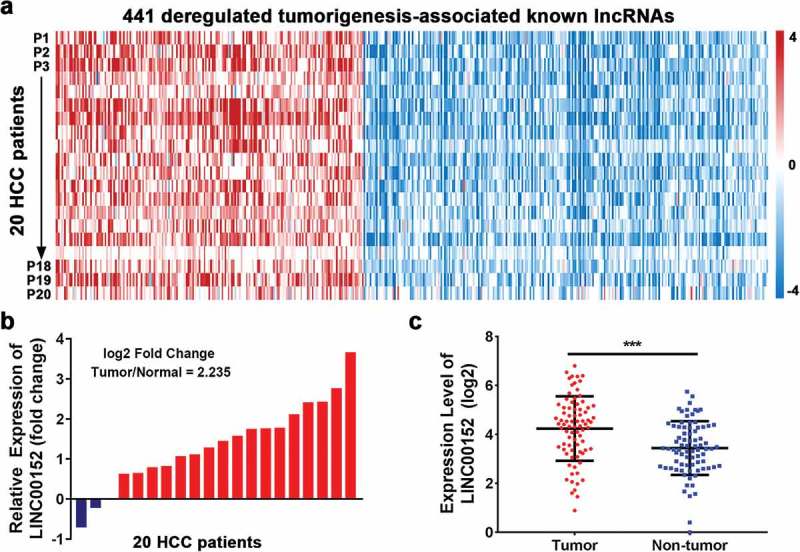
Recurrently dysregulated lncRNAs and LINC00152 expression in HCC tissues. (a) Analysis of 441 recurrently dysregulated tumourigenesis-associated lncRNAs according to the NCBI Gene Expression Omnibus bioinformatics tool (GEO Series Accession Number GSE77509). lncRNAs were rank-ordered by differential expression between adjacent normal tissue and primary tumour samples (n = 20). (b) Relative expression of LINC00152 in HCC compared with adjacent non-neoplastic liver tissues according to NCBI Gene Expression Omnibus analysis. The fold change was ~2.24 higher in tumour than in normal tissues (n = 20). (c) Upregulation of LINC00152 in HCC tissues compared with non-tumour tissues as analysed by qRT-PCR. LINC00152 expression was normalized to that of U6 (n = 80) (*** p < 0.001).
