Figure 2.
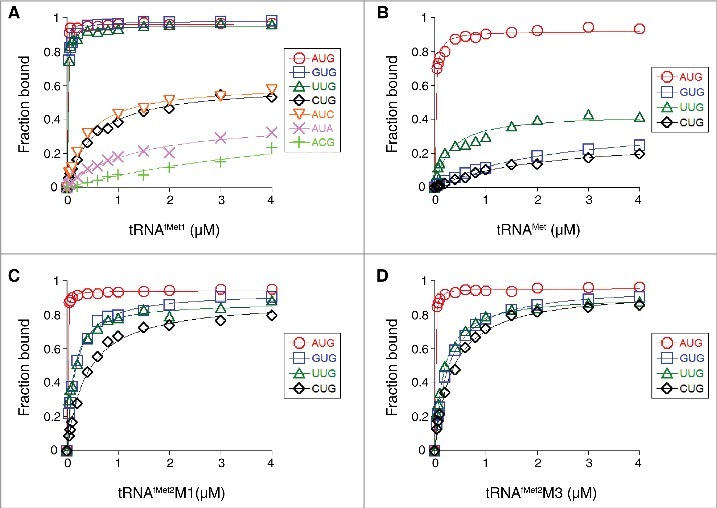
Effects of the start codon and tRNA sequence on the thermodynamic stability of the 30S•mRNA•tRNA complex. 30S subunits (1 µM) were incubated with mRNA (0.01 µM; with indicated start codon and preannealed radiolabeled primer) and various concentrations of tRNAfMet1 (A), tRNAMet (B), tRNAfMet2M1 (C), or tRNAfMet2M3 (D) for 2 h at 37°C, and then complexes were analyzed by toeprinting. Fraction of bound mRNA (F) was quantified as [toeprint signal / (toeprint + run-off signal)] and plotted versus input tRNA concentration. Data were fit to the equation F = Fmax[bc/(bc+1/K TC)], where b is the input tRNA concentration, c is the input 30S concentration, K TC is the equilibrium association constant, and Fmax is the maximal level of detected complex. For the ACG case of panel A, the Fmax parameter was set arbitrarily at 1.0 prior to fitting the curve shown. All K TC and Fmax values are listed in Table 1.
