Figure 1.
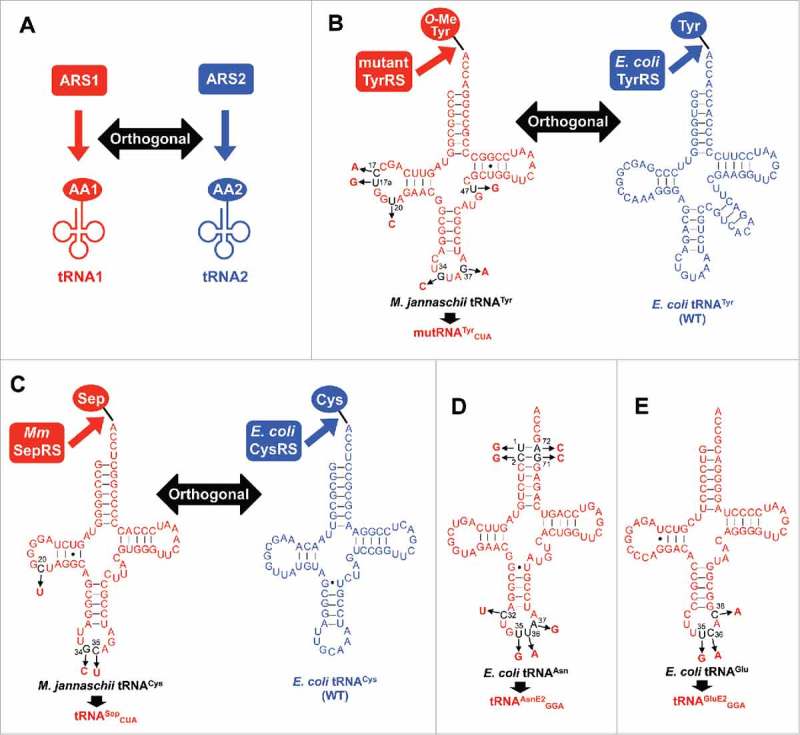
Development of orthogonal tRNAs. (A) Conceptual scheme of orthogonal ARS/tRNA pairs. ARS1 specifically charges amino acid 1 (AA1) onto tRNA1, but not onto tRNA2. Conversely, tRNA2 is exclusively charged with amino acid 2 (AA2) by ARS2, but not with AA1 by ARS1. (B) Mutant TyrRS/tRNATyr CUA pair orthogonal to the E. coli wild-type (WT) TyrRS/tRNATyr pair. The mutant TyrRS was developed based on M. jannaschii TyrRS to charge O-methyltyrosine instead of tyrosine on M. jannaschii tRNATyr or mutRNATyr CUA. The mutRNATyr CUA has 5 nucleotide substitutions, C17A, U17aG, U20C, G37A, and U47G, to improve the orthogonality as well as the anticodon substitution from GUA to CUA to decode UAG codon. (C) Methanococcus maripaludis (Mm) SepRS/tRNASep CUA pair orthogonal to the E. coli wild-type (WT) CysRS/tRNACys pair. The tRNASep CUA was designed based on M. jannaschii tRNACys with 3 nucleotide changes (C20U, G34C, and C35U). (D) Secondary structure of tRNAAsnE2 GGA designed based on E. coli tRNAAsn. Nucleotide changes at the acceptor stem are introduced to give orthogonality to the other E. coli ARS/tRNA pairs. (E) Secondary structure of tRNAGluE2 GGA designed based on E. coli tRNAGlu.
