Fig. 7.
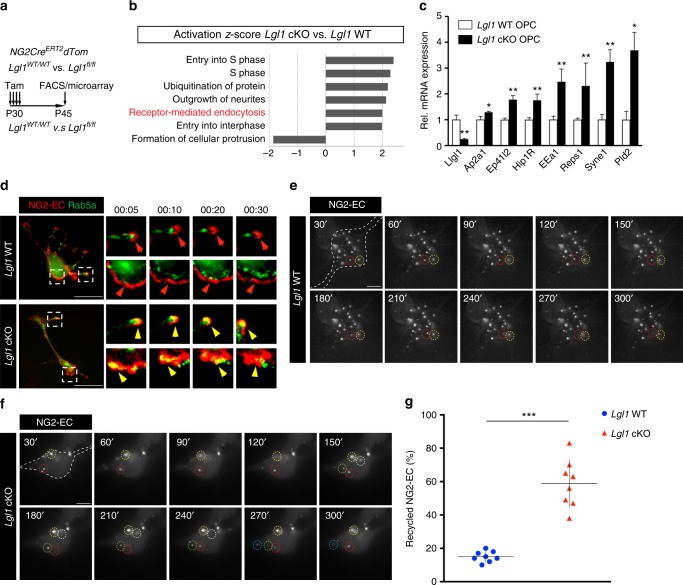
Lgl1 negatively regulates internalization and recycling of NG2 in OPC. a Schematic illustration of the experimental approach assessing global changes in gene expression by microarray in Lgl1 WT vs. Lgl1 cKO OPC. FACS sorted red fluorescent PDGFRα+O4− cells. b Bar graph of ingenuity pathway analyses (IPA) of Lgl1 WT vs. cKO OPC. Receptor-mediated endocytosis pathway is amongst the upregulated pathways in Lgl1 cKO cells. c Validation by q-RT-PCR for mRNA encoding receptor-mediated endocytosis regulators that were differentially expressed in Lgl1 cKO cells as determined by microarray analyses. OPC were derived from the corpus callosum of GFAP-Cre, Tom, Lgl1wt/wt and, GFAP-Cre, Tom, Lgl1fl/fl mice. Data represented as mean ±s.e.m. n = 6 Lgl1 WT and 6 Lgl1 cKO animals (*p < 0.05, **p < 0.01, Mann–Whitney test). d Steady-state frames of time lapse (1 frame/ 10 min) of NG2-EC (red) and early-endosome marker Rab5a (green) in Lgl1 cKO and WT OPC. Note that NG2 stays mostly at the membrane in Lgl1 WT cells (red arrows), whereas it co-localizes with the early endosome in the cytoplasm of Lgl1 cKO OPC (yellow arrows). Scale bar: 20 µm. e, f Frames illustrating time lapse experiment under TIRF microscopy. Pictures are acquired each 30 seconds as soon as cells were exposed to differentiation conditions. Each white puncta in focus represents labeled NG2-EC docked at the membrane. Individual recycled puncta are highlighted with dashed circles of different colors. Note that in Lgl1 cKO cells, the majority of the labeled NG2 is internalized and recycled to the membrane whereas in WT cells, the majority of NG2 puncta retains a stable localization at the membrane. Scale bars: 10 μm. g Graph representing the quantification of NG2 recycling at the membrane throughout the first 5 min of differentiation conditions. Data represented as mean ± s.e.m. n = 3 cells per genotype, three independent experiments (***p < 0.005, Student's t-test)