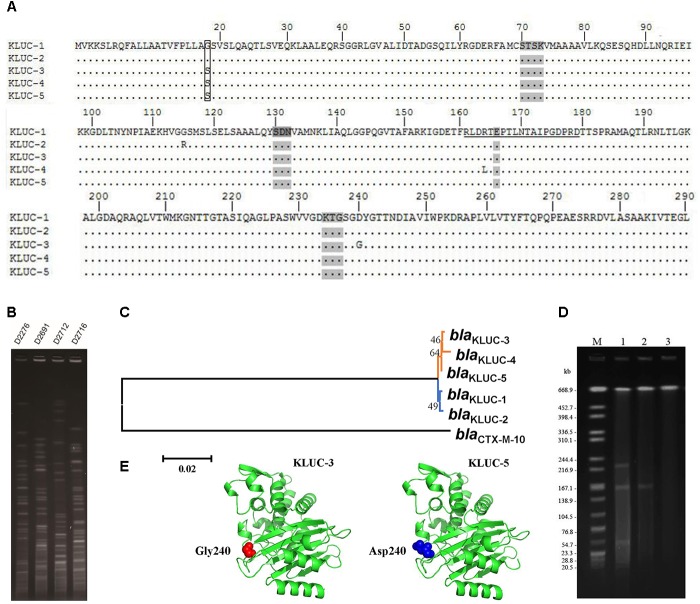
FIGURE 1.
Sequence comparison, transferability and phylogenetic analyses of blaKLUC-5. (A) Alignment of KLUC-5 with four other KLUC subtypes. Amino acids are numbered according to the standard numbering scheme for the class A β-lactamases. Ellipses indicate identical amino acid residues. The amino acids composing the omega loop are underlined. Four structural elements characteristic of class A β-lactamases are shaded (70SXXK73, 130SDN132, 166E and 234KTG236). The substituted amino acids between KLUC-5 and KLUC-1 are boxed. The four KLUC subtypes, KLUC-1, KLUC-2, KLUC-3, and KLUC-4, were retrieved from the NCBI under accession numbers AAK08976, ABM73648, JX185316, and JX185317, respectively. (B) PFGE analysis of four blaKLUC-harboring E. coli clinical isolates. (C) Phylogenetic analysis of five blaKLUC genes. Branches with the same color indicate that the genes were isolated from the same district. (D) S1-PFGE patterns of KP1276, transconjugant, and EC600. Lane M is the size marker strain Salmonella serotype Braenderup H9812 digested with XbaI. Lanes 1–3 are the genomes of strain KP1276, transconjugant, and EC600 digested with S1 Nuclease. (E) The three-dimensional structure of KLUC-3 and KLUC-5. The atoms belonging to amino acid at position 240 of KLUC-3 and KLUC-5 are indicated.