Abstract
Background
Genomic testing is often limited by the exhaustible nature of human tissue and blood samples. Here we describe biotinylated amplicon sequencing (BAmSeq), a method that allows for the creation of PCR amplicon based next-generation sequencing (NGS) libraries while retaining the original source DNA.
Design and methods
Biotinylated primers for different loci were designed to create NGS libraries using human genomic DNA from cell lines, plasma, and formalin-fixed paraffin embedded (FFPE) tissues using the BAmSeq protocol. DNA from the original template used for each BAmSeq library was recovered after separation with streptavidin magnetic beads. The recovered DNA was then used for end-point, quantitative and droplet digital PCR (ddPCR) as well as NGS using a cancer gene panel.
Results
Recovered DNA was analyzed and compared to the original DNA after one or two rounds of BAmSeq. Recovered DNA revealed comparable genomic distributions and mutational allelic frequencies when compared to original source DNA. Sufficient quantities of recovered DNA after BAmSeq were obtained, allowing for additional downstream applications.
Conclusions
We demonstrate that BAmSeq allows original DNA template to be recovered with comparable quality and quantity to the source DNA. This recovered DNA is suitable for many downstream applications and may prevent sample exhaustion, especially when DNA quantity or source material is limiting.
Abbreviations: BAmSeq, Biotinylated amplicon sequencing; NGS, Next generation sequencing; FFPE, Formalin-fixed paraffin embedded; cfDNA, Circulating cell-free DNA; gDNA, Genomic DNA; pDNA, Plasma DNA; qPCR, Quantitative polymerase chain reaction; ddPCR, Droplet digital PCR
Keywords: Next generation sequencing, Plasma DNA, Droplet digital PCR (ddPCR), Targeted amplicon sequencing
Highlights
-
•
Modification of targeted panel sequencing allows for recovery of original DNA template.
-
•
Protocol provides value in the setting of scarce DNA template.
-
•
Recovered DNA is suitable for NGS, ddPCR and qPCR.
-
•
Recovered DNA shows no loss of genomic regions.
1. Introduction
The use of patient samples for cancer research has led to the development of new diagnostics and therapeutics for the treatment of human cancers. In particular, tissue specimens have been essential for understanding the mutational landscapes of cancer [1], and DNA extraction from primary and metastatic tumors is now routine practice [2]. Similarly, circulating cell free DNA (cfDNA) or “liquid biopsy” is an emerging analyte for many applications in clinical oncology and cancer research [3], [4], [5], [6]. The utility of cfDNA for cancer mutation detection with digital PCR and NGS has been shown for a number of clinically relevant oncologic issues including response to therapy [7], [8], detection of driver and resistance mutations [9], [10] and measurement of residual disease burden [11]. Unfortunately, a limitation of both tissue samples and cfDNA is their exhaustible nature [12], limiting the number of tests and assays that can be performed. This is especially relevant when performing NGS on a limited number of loci, where often the entire sample is used to maximize the chance of detecting a rare mutation within many wild type DNA molecules.
Techniques such as Safe-SeqS [13] and Duplex Sequencing [14] have been developed for increasing the sensitivity of targeted amplicon sequencing [15]. These techniques incorporate barcoding strategies to distinguish PCR and NGS errors from true rare mutant alleles. However, the ultimate sensitivity of these methods depends on the starting amount of DNA, often necessitating the use of the entire sample. To address this problem, we describe a novel modification for amplicon NGS termed Biotin Amplicon Sequencing or BAmSeq. This modification of amplicon sequencing enables recovery of the sample DNA template for future use in other molecular assays, and importantly does not compromise the quantitative integrity of the original source DNA. The use of BAmSeq allow for recycling of DNA for samples of limited quantities.
2. Materials and methods
2.1. BAmSeq
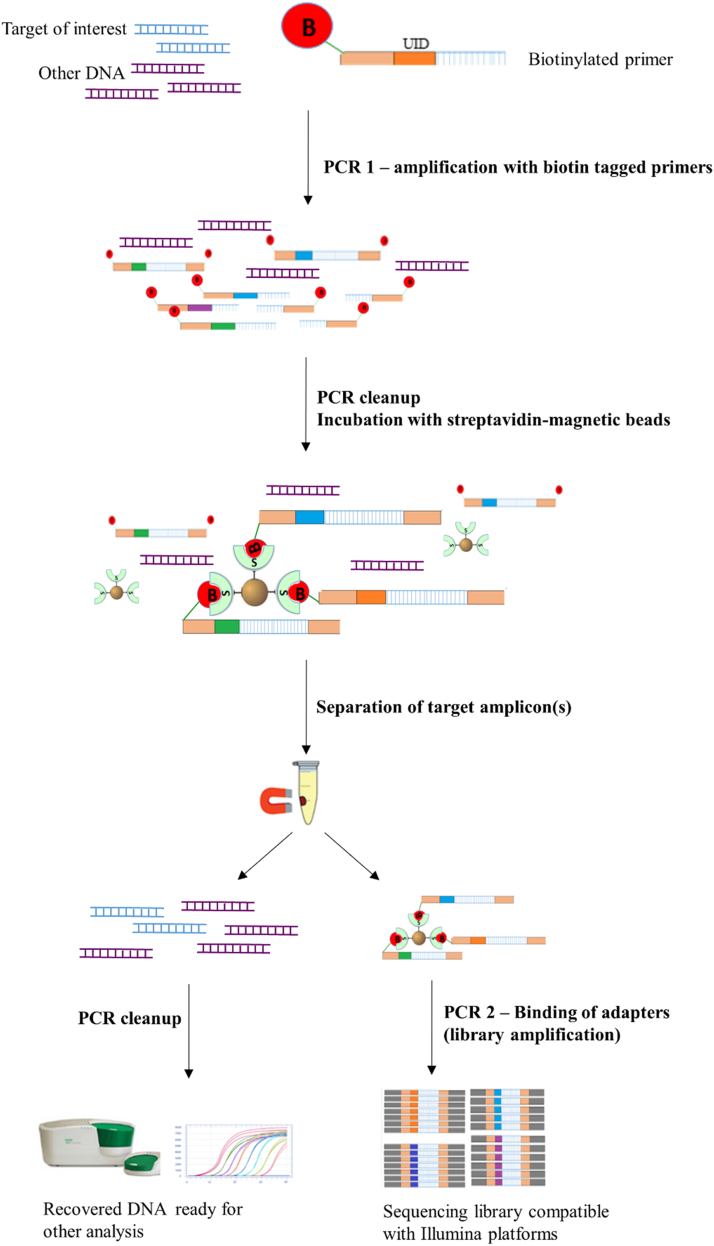
A 5’ biotin molecule and subsequent streptavidin pull down step was added to the overall methodology of the Safe-SeqS technique [13]. Safe-SeqS was developed to increase the sensitivity of mutation detection by tagging amplicons of interest using a random sequence of 12–14 nucleotides called “unique identifiers”. The protocol barcodes DNA strands during the first PCR step followed by a secondary PCR in which Illumina adapter sequences are added to the tagged template. In the BAmSeq approach described in Fig. 1, first, a 5’ biotin molecule is attached to the amplicon of interest by using biotinylated forward and reverse primers during the first PCR step. PCR reactions were performed in a 50 µL final volume with final concentrations of 1x Phusion® HF Buffer, 200 µM dNTP's, 5% DMSO, 0.5 µM forward and reverse primers, and 2 units of Phusion® Hot Start Polymerase (New England Biolabs) using the following cycling conditions: an initial denaturation at 98 °C for 30 s (s), followed by one cycle of 98 °C for 10 s, 64 °C for 15 s, and 72 °C for 30 s. Subsequent cycles (2–5 depending on the amplicon) maintained the same temperatures and times for denaturation and elongation steps, while the annealing temperature was changed from 64 °C to 61 °C. Upon completion of the first PCR step, the PCR products are now covalently linked to biotin molecules. Second, the sample was cleaned using AMPure XP PCR Purification magnetic beads (Agencourt) per the manufacturer's recommendation. Third, the sample was eluted into 20 µL of water and incubated in equal volume of Dynabeads® MyOne™ Streptavidin (ThermoFisher Scientific) to extract all the biotin-tagged DNA strands, including residual primers. Prior to the incubation, Streptavidin beads were washed to remove buffers and preservatives as specified in the manufacturer's protocol. An equal volume of cleaned streptavidin beads was added to the eluent and incubated for at least 1 h at room temperature with mild shaking to maintain the beads in suspension. After incubation, the tubes were placed in a magnetic stand to separate the supernatant (containing the original source DNA) and the magnetic beads (bound to amplicon of interest), until the solution appears clear (10–15 min). Subsequently, the two samples collected are:1) the supernatant containing the recovered DNA and residual buffers. This sample was carefully removed and collected in a microcentrifuge tube, cleaned using AMPure XP beads, and either stored at − 20 °C, or used for further applications, and 2) the streptavidin-biotin-amplicon complex. The sample was washed twice with 1X B&Q buffer and once with distilled water, then resuspended in 22 µL of water as per the manufacturer's recommendations.
Fig. 1.
Schematic representation of BAmSeq protocol. Procedural outline for BAmSeq protocol described in this study. The biotinylated primers were similar to [13] with the addition of biotin at the 5’ end. Primers for PCR 2 (library amplification) are included in Illumina's TruSeq DNA library preparation kits. (Note: inclusion of “Unique Identifier” is not required for BAmSeq).
The eluted streptavidin-biotin-DNA complex beads are used as template for the second PCR step, in which Illumina-specific adapters are added, as well as indexes (unique 6 bases) for multiplexing. Because the entire complex can be used directly as template for the PCR reaction, denaturation of the streptavidin-biotin complex is not necessary. The following thermocycling conditions were used for this amplification step: 25–35 cycles (depending on initial DNA concentration used) at 98 °C for 10 s (s), 66 °C for 30 s, and 72 °C for 40 s. Upon PCR conclusion, the entire reaction is placed on a magnet and allowed to separate (10–15 min). Without disturbing the pellet, the supernatant (sequencing library) is collected, and the magnetic beads stored at − 20 °C or discarded. The newly isolated sequencing library then undergoes an additional clean-up step using the AMPure XP beads. Cleaning is followed by quantification using the KAPA Library quantification kit, a qPCR assay by Bio-Rad®, and subjected to NGS with the Illumina platform. A schematic of the protocol is shown in Fig. 1, and a complete list of all primers used in this study can be found in Supplemental Table S1.
2.2. Patient and sample collection
All patients were consented and enrolled in an IRB protocol at the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center (JHSKCCC; Baltimore, MD) approved for collection and genomic analysis of tissue and bodily fluids from breast cancer patients for use in research. FFPE normal tissue samples and plasma DNA from four patients (two patients with breast cancer and two normal plasma samples) were collected and used for the study.
2.3. Cell culture for genomic DNA isolation
Cell lines were chosen for the presence or absence of a heterozygous variant in regions of interest. HCT-15 is a human colorectal cell line that harbours a D144G variant in exon 7 of the SPOP gene, U87MG is a human brain cancer cell line that harbours a C228T mutation in the TERT promoter region, while MCF10A is a human mammary epithelial cell line that is wild type for both specified loci. The non-transformed human epithelial cell line MCF10A was purchased from ATCC and cultured in DMEM/F12 (1:1) supplemented with 5% horse serum (Life Technologies, Carlsbad, CA), 20 ng/mL epidermal growth factor (Sigma-Aldrich, St. Louis, MO), 0.5 µg/mL hydrocortisone (Sigma-Aldrich), 10 µg/mL insulin (Life Technologies), penicillin-streptomycin (Life Technology), and 0.1 µg/mL cholera toxin (Sigma-Aldrich). The U-87 MG brain cancer cell line was maintained in DMEM media with 10% fetal bovine serum (Life Technologies), and 1% of penicillin-streptomycin (Life Technology). HCT-15 colorectal cells were maintained in RPMI-1640 basal medium with 10% fetal bovine serum (Life Technologies) and 1% of penicillin-streptomycin (Life Technology). Genomic DNA for all cell lines was prepared using the QIAamp DNA Blood Mini Kit following the manufacturer's protocol. DNA concentration was calculated using the Quant-iT™ PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, Waltham, MA).
2.4. Formalin-fixed paraffin-embedded (FFPE) tissue and plasma DNA collection
FFPE tissue DNA was isolated using the GeneRead DNA FFPE kit from Qiagen, while plasma DNA was extracted as described [11] using the QIAamp Circulating Nucleic Acid kit (Qiagen).
2.5. Cancer panel amplicon sequencing
We utilized the Swift Bioscience ACCEL Amplicon Panel which provides coverage of 263 amplicons targeting 56 cancer associated genes. The targeted library is compatible with fragmented DNA samples including FFPE DNA and plasma DNA, which can be sequenced using any Illumina platform. For our study, all libraries were prepared and sequenced using an Illumina MiSeq following the manufacturer's specifications including quantification by a PCR based assay (KAPA Library Quantification Kit, KAPA BioSystem). The total length of reads was 138 bp on average based on the manufacturer's website. The library was loaded at concentrations ranging from 10 pM to 16 pM, and exact amount was determined based on the desired cluster density and MiSeq reagent kit utilized.
2.6. Quantitative PCR (qPCR) and droplet digital PCR
qPCR was carried out using SYBR Green based detection with an iCycler (Bio-Rad) following a protocol as previously described [16]. Droplet digital PCR (ddPCR) (Bio-Rad) was performed as per the manufacturer's recommendation using an EvaGreen Supermix assay. The fluorescence per droplet is assessed, and the fraction of positive to negative droplets was recorded using the QuantaSoft software. The experiment was set up as “ABS analysis” which allows for automatic threshold determination by the QuantaSoft software, yielding absolute quantification of a target amplicon as (copies/µL). Because this study focused solely on comparing the presence of DNA template in pre- versus post- BAmSeq treated DNA, no pre-amplification was conducted prior to ddPCR, each target was assayed in triplicate independent wells and all replicates contained> 10,000 droplets. Primers for all PCR amplicons are listed in Supplemental Table S1.
2.7. Sequencing data analysis
Raw sequencing data was used to assess and track the quality of the sequencing data. Raw data passing the quality controls was further analyzed by aligning to Human Genome (build GRCh37.p13/hg19) [17] reference using BWA aligner [18] (v0.7.10). Post alignment data was passed through Picard Tools (V1.125) [19] to assess the alignment quality. Aligned data that passed quality control was used to call the variants using an in-house variant caller, MDLVC, which scans through the alignment data for raw variant calls. Resulting raw variants were further filtered with various parameters including a minimum base quality of Q25, minimum base depth of 1000, and strand bias filters. In addition to the above filters, false positive variant calls were assessed, tracked and filtered out using negative controls sequenced with this study. Variant calls that code for silent changes or that are designated as frequent in populations by dbSNP [20], ExAC [21] TCGA [22] and ClinVar [23] reference databases were excluded from analysis. High confidence somatic variants were corroborated with mutant allele frequency, and tumor cellularity. For analysis of plasma DNA, variants allelic frequencies below 1% were filtered out to minimize potential false positives. Additionally, variants were annotated with information from COSMIC [24] and mutation impact annotation from Mutation Assessor Impact prediction [25]. Final variant calls were visualized and further validated using Integrated Genome Viewer, IGV [26].
3. Results
3.1. The addition of biotin molecules to NGS primers does not affect sequencing
To determine whether the addition of biotin molecules would negatively impact NGS library preparation, we created targeted amplicon sequencing libraries using the BAmSeq method. Samples consisted of digested genomic DNA from three cell lines: MCF10A, HCT-15 and U87MG. Cell lines were chosen based on the presence or absence of a heterozygous variant in regions of interest. HCT-15 is a human colorectal cell line that harbours a c.431 A>G variant (p.D144G) on exon 7 of the SPOP gene, U87MG is a human brain cancer cell line that harbours a g.228 C>T TERT promoter region mutation and MCF10A a human mammary epithelial cell line which is wild type for both loci. Prior to use for BAmSeq, Sanger was used to confirm the presence or absence of variants in cell lines (Supplemental Fig. S1). For these experiments, DNA was isolated, quantified and mixed in various combinations to create samples with user-specified mutant to wild type frequencies. Samples labeled S1–S5 refer to combinations of MCF10A and HCT-15, while T1-T5 correspond to MCF10A-U87MG mixtures, which were then used for BAmSeq (Supplemental Fig. S2A). The BAmSeq method created targeted amplicon sequencing libraries for analysis that were quantified and loaded into a MiSeq Illumina platform for NGS. All prepared libraries were successfully equenced, demonstrating that the addition of a 5’ biotin molecule to primers or subsequent streptavidin pull down steps had no effect on the generation and use of NGS libraries. To determine if there was any amplification bias during the streptavidin template separation, we compared the expected mutant percentages to the allele frequencies obtained after sequencing. FASTQ files were aligned using Bowtie [27] and the allelic frequencies determined by visually analyzing the aligned data using the Integrative Genomics Viewer [26], [28]. NGS results showed that the variant/mutant percentages observed were consistent with the expected percentages as summarized in Table 1.
Table 1.
Expected and observed mutation frequencies after NGS of libraries generated by BAmSeq.
| Sample ID | DNA source | Expected mutant allelic frequency % | Observed mutant allelic frequency % |
|---|---|---|---|
| S1 | gDNA (HCT15 + MCF10A) | 50% | 48% |
| S2 | gDNA (HCT15 + MCF10A) | 25% | 25% |
| S3 | gDNA (HCT15 + MCF10A) | 10% | 13% |
| S4 | gDNA (HCT15 + MCF10A) | 5% | 7% |
| S5 | gDNA (HCT15 + MCF10A) | 3% | 3% |
| T1 | gDNA (U87GM + MCF10A) | 50% | 47% |
| T2 | gDNA (U87GM + MCF10A) | 25% | 21% |
| T3 | gDNA (U87GM + MCF10A) | 15% | 15% |
| T4 | gDNA (U87GM + MCF10A) | 10% | 12% |
| T5 | gDNA (U87GM + MCF10A) | 5% | 6% |
| T11 | Recovered DNA from sample T1 | 50% | 53% |
| T12 | Recovered DNA from sample T2 | 25% | 24% |
Column one: S = SPOP libraries; T = TERT libraries. Column two: source of genomic DNA (gDNA). Column three: expected mutation percentage based on user specified wild type (WT) to mutant ratios. Column four: allelic frequency obtained during sequencing.
3.2. Recovered DNA retains relative copy number of source DNA
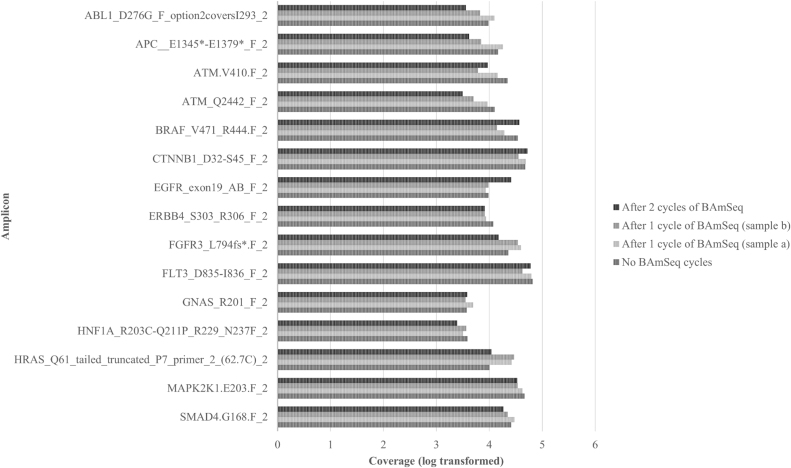
We initially assessed whether BAmSeq had any overt effects on relative copy number by comparing samples before and after BAmSeq treatment. To determine this, we used a cancer gene amplicon panel to create standard sequencing libraries from plasma DNA as well as recovered DNA after BAmSeq. Plasma DNA was separated into four distinct aliquots. One aliquot was left untreated as a control labeled “Before”, referring to the original source DNA prior to BAmSeq. Two separate aliquots were subjected to one round of BAmSeq and served as technical duplicates. This DNA was recovered and labeled “After-1a” and “After-1b”. The last aliquot was put through two subsequent rounds of BAmSeq and labeled “After-2” (Supplemental Fig. S3). NGS libraries were created from each recovered DNA sample and sequenced. Fifteen randomly chosen amplicons with their respective total coverage (log transformed) are found in Fig. 2, and coverage data for all amplicons in the panel can be found in Supplemental Fig. S4. Results of total coverage per amplicon show no observable loss of genomic regions when comparing untreated versus recovered DNA after one or two rounds of BAmSeq (Fig. 2). Samples receiving one round of BAmSeq (After-1a and After-1b) were performed in parallel and produced comparable levels of amplicon coverage. Two independent experiments were conducted using plasma DNA with concordant results.
Fig. 2.
Sequencing coverage (bars) for recovered plasma DNA. Sequencing libraries were prepared using a cancer gene panel targeting 263 loci. Samples were recovered DNA following zero (No BAmSeq cycle), one (After 1), or two (After-2) cycles of BAmSeq. Coverage for 15 amplicon subset randomly selected and displayed above (See Supplemental S4 for all amplicon coverage).
3.3. Recovered DNA retains allelic frequency
After determining that copy numbers are relatively well preserved in recovered DNA, we investigated whether allelic frequencies were also retained. First, we used recovered DNA from samples T1 and T2 which contained genomic DNA artificially mixed to mutant allelic frequencies of 50% and 25%, respectively. The recovered DNA was processed through BAmSeq a second time targeting the same amplicons (TERT promoter) and the newly created sequencing libraries labeled T11 and T12 (Supplemental Fig. S2B). Sequencing results suggest that the mutant allelic frequencies were retained for both T11 and T12 with estimated frequencies of 53% and 24%, respectively (Table 1). Next, we wanted to determine if similar results would be observed using plasma DNA as starting material. Using the NGS data generated from our plasma DNA BAmSeq libraries to evaluate copy number (Fig. 2 and Supplemental Fig. S4), we applied a proprietary pipeline for aligning, filtering and variant calling as described in the Methods section. For plasma DNA from a healthy donor, a total of five commonly reported polymorphisms were detected in all samples at comparable allelic frequencies as shown in Table 2.
Table 2.
Variant allelic frequencies for benign SNP's detected in recovered plasma DNA samples after BAmSeq.
| Variant Allelic Frequency (%) per sample |
|||||
|---|---|---|---|---|---|
| Gene | AA change | Before BAmSeq plasma DNA | After (1) BAmSeq plasma DNA 1a | After (1) BAmSeq plasma DNA 1b | After (2) BAmSeq plasma DNA |
| KDR | p.Q472H | 51.16 | 49.227 | 52.53 | 57.16 |
| RET | p.L769L | 49.35 | 49.783 | 49.85 | 49.48 |
| ATM | p.A1931A | 50.6 | 50.7 | 50.70 | 50.18 |
| FGFR3 | p.T539T | 99.59 | 99.573 | 99.11 | 99.82 |
| PDGFRA | p.P567P | 99.66 | 99.578 | 98.97 | 99.82 |
Samples of plasma DNA were control (“Before” BAmSeq) or recovered DNA collected after one (“After 1”) or two (“After 2”) rounds of BAmSeq. “After 1” recovered DNA was processed in duplicate and all samples were aliquoted (columns under allelic frequency) from the same source DNA (rows).
Lastly, the same set of experiments was performed with a sample of plasma tumor DNA taken from a patient with breast cancer to detect potential mutations at lower allelic frequencies. All mutations identified in the “Before” samples were also detected in the “After” samples, with the lowest allelic frequency reported being 1.07%. Although variants with allelic frequencies< 1% were obtained during the analysis, a threshold of at least 1% was applied to all sequencing data to minimize background and possible false positives arising from the sequencing process. Two duplicates were prepared for each BAmSeq procedure to account for potential variation between samples, all allelic frequencies detected are summarized in Table 3.
Table 3.
Allelic frequencies for mutations detected in recovered plasma tumor DNA samples before and after BAmSeq.
| Variant allelic frequency (%) per sample |
|||||||
|---|---|---|---|---|---|---|---|
|
Before BAmSeq |
After (1) BAmSeq |
After(2) BAmSeq |
|||||
| Gene | AA change | 1 | 2 | 3 | 4 | 5 | 6 |
| MSH6 | p.F1088fs*5 | 1.22 | 1.87 | 4.76 | 3.45 | 1.97 | 1.07 |
| PIK3CA | p.E545K | 1.07 | 2.17 | 4.25 | 1.43 | 3.58 | 3.27 |
| MLH1 | p.R385C | 4.35 | 8.60 | 6.92 | 8.12 | 10.30 | 16.68 |
| EGFR | p.S768I | 7.22 | 6.62 | 10.09 | 11.82 | 9.37 | 12.64 |
| FGFR1 | p.D133delD | 2.87 | 3.92 | 5.35 | 2.76 | 2.73 | 3.74 |
Three treatment samples (in duplicate) included: an untreated control (“Before” BAmSeq) and two treated samples that underwent (1) or (2) rounds of BAmSeq denoted as “After (1)” or “After (2)” respectively. Duplicates are labeled as “1” and “2” for the untreated controls, “3” and “4” for samples put through one round of BAmSeq and “5” and “6” for samples treated with two subsequent rounds of BAmSeq.
3.4. Recovered-DNA can be used for different molecular assays
Recovered DNA from several samples was tested to determine if the DNA could be used for analyses such as endpoint, quantitative, and ddPCR. Several DNA sources were utilized for each assay, and recovered DNA following BAmSeq was compared to the corresponding untreated DNA source as detailed in Supplemental Fig. S5.
3.4.1. End-Point PCR
Recovered DNA samples were subjected to end-point PCR for five amplicons (e.g., HER2 exon 19, SPOP exon 6, SPOP exon 7, PIK3CA exon 29, and ESR1 exon 10) to assess if the template was suitable for amplification. Supplemental Fig. S6 contains images from nine recovered DNA samples. Three independent groups labeled set 1, 2 and 3 contained recovered DNA originating from genomic (“g”), plasma (“p”) and FFPE (“f”) sources. All were queried for the presence of five amplicons of interest and positive bands (~ 100 bp) were detected in all.
3.4.2. Quantitative&droplet digital PCR
To determine if recovered DNA was compatible with quantitative assays such as qPCR and ddPCR, we queried for the presence of amplicons in “Before” samples (DNA not used in BAmSeq), and “After” samples (recovered DNA after one round of BAmSeq) as detailed in Supplemental Fig. S5. For qPCR, quantification cycle (Cq) values for each replicate were obtained as an indirect measure of DNA concentration per amplicon. Similar Ct values were obtained for all samples per amplicons tested (e.g., SMAD, SBNO2, P2RY2, and ABCA3) (Fig. 3)
Fig. 3.
Quantitative and droplet digital PCR using recovered DNA. Bars depict cycle threshold values (Ct; y-axis) obtained from qPCR analysis for three independent DNA samples (x-axis) compared to control (“Before” BAmSeq).
Additionally, we used ddPCR to demonstrate absolute copy number of amplicons of interest, “Before” and “After” BAmSeq. Fig. 4 displays a representative sample depicting positive droplets per amplicon for “After” and corresponding “Before” reference. The calculated concentration (copies/µL) and 95% confidence interval per amplicon for all tested samples was determined by QuantaSoft software and displayed in Supplemental Table S2. The differences between observed and expected values for both qPCR and ddPCR assays were minimal.
Fig. 4.
Quantitative and droplet digital PCR using recovered DNA. Positive PCR droplets and total number of events (bars; y-axis) versus amplicons (four; x-axis) with the top figure depicting “Before” BAmSeq control and the bottom figure depicting recovered “After” BAmSeq.
4. Discussion
Although newer technologies such as ddPCR and NGS have allowed for an unprecedented analysis of the human genome, analysis is often limited by the exhaustible nature of tissue and blood samples. This is particularly relevant for oncology where rare mutation detection in plasma DNA or a tumor biopsy mandates assessing the maximum number of genome equivalents for optimal sensitivity. This can exhaust a single sample and limits further downstream molecular analyses. Here, we provide a simple modification to targeted sequencing called BAmSeq, which allows for the recovery of the original DNA material that is suitable for additional analyses. The BAmSeq modification requires the addition of biotin primers with a subsequent streptavidin pull down step during NGS library preparation. Our modifications had minimal effect on NGS of recovered DNA libraries as highlighted in Table 1. Subsequent analysis of BAmSeq DNA also demonstrated that DNA copy number and allelic fractions in recovered DNA after one and two procedures were comparable to starting DNA material (Table 2). Importantly, these results appeared to be consistent across different preparations of DNA including cell line genomic DNA, plasma DNA and FFPE DNA derived from patient samples.
By subjecting the recovered DNA to a cancer sequencing panel targeting 263 amplicons, we determined that the copy numbers across these loci were retained (Fig. 2 and Supplemental Fig. S4). These observations suggest that the recovered DNA's genomic landscape is relatively unchanged, with minimal losses or gains of the genetic loci after the BAmSeq protocol. This may be beneficial when it is desirable to analyze a patient sample through more than one round of sequencing or when using different targeted panels sequentially [29]. Furthermore, we showed that the allelic frequency within NGS libraries created from recovered DNA was conserved when looking at homozygous and heterozygous variants before and after BAmSeq.
To ensure that recovered DNA after BAmSeq was usable in downstream assays, we performed commonly used techniques beyond NGS, namely qPCR and ddPCR. These technologies are becoming increasingly important for quantification of clinical and research samples in cancer research and other biomedical fields [9], [11], [30], [31]. Given that these assays are known to be negatively affected by the presence of inhibitors [32] we wanted to verify that the BAmSeq protocol would not adversely affect the ability to use recovered DNA for these purposes. Using a variety of template DNA sources, as well as recovered DNA from one and two BAmSeq procedures, we showed that the recovered DNA remained a suitable template for downstream molecular analyses.
It is worth noting that every subsequent round of BAmSeq will see some loss of DNA. This is inevitable as recovery of DNA can never be 100%. However, we found that the amount of DNA lost during the BAmSeq procedure is greatly influenced by the careful recovery of the supernatant during the streptavidin-biotin separation step. Thus, meticulous pipetting of the recovered DNA after pull-down is key to maximizing DNA yield.
An argument could be made that use of genome wide amplification kits, or the remaining NGS library could be used for downstream analyses. However, a potential concern with these approaches is the possible amplification bias in terms of copy number, as well as artifactual mutations from the PCR amplification used for the majority of these techniques. This is especially relevant when assaying for rare tumor mutations in plasma DNA, where even high fidelity polymerases can lead to artifacts [33]. Artifacts can affect downstream assays by skewing allelic fractions, introducing false mutations, and affecting the performance of molecular probes [34], [35]. Furthermore, BAmSeq provides the capability of using the same sample to increase technical replicates for sequencing. It has been reported that increasing technical replicates and not sequencing depth is more effective at decreasing error rates, a key consideration when using NGS technology for diagnostic purposes [36]. Additionally, stochastic variability linked with variant callers, library preparation and inherent variability during sequencing (e.g., quantification, loading and base calling) can be minimized by including additional replicates, which increases the utility of the BAmSeq protocol.
In summary, we describe a method for extending the usability of exhaustible DNA sources for NGS and various PCR techniques. The ability to use DNA for sequential testing and analysis may prove useful for a number of clinical and research assays. In particular, the application of BAmSeq to cancer genomic studies using FFPE and plasma DNA may allow for orthogonal cross validation of mutations and/or copy number alterations within a given sample. Importantly, BAmSeq will allow for the maximum utilization of precious and exhaustible patient samples and provides a more complete and accurate analysis with current and future technologies.
Acknowledgments
None.
Acknowledgments
Author contributions
All authors had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Conception and design: KC, AM, BHP
Development of methodology: All authors
Acquisition of data: KC, AM, AP
Analysis and interpretation of data: All authors
Writing, review, and/or revision of the manuscript: All authors.
Acknowledgments
Conflict of interest disclosures
B.H.P. has ownership interest and is a paid member of the scientific advisory board of Loxo Oncology and is a paid consultant for Foundation Medicine, Inc and H3 Biomedicine. Under separate licensing agreements between Horizon Discovery, LTD and The Johns Hopkins University, B.H.P. is entitled to a share of royalties received by the University on sales of products. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. All other authors declare no potential conflicts.
Acknowledgments
Funding/support
This work was supported by: The Komen Foundation (B.H.P.), NIH CA088843 and CA194024 (K.C. and B.H.P.), We would also like to thank and acknowledge the support of the Sandy Garcia Charitable Foundation, the Commonwealth Foundation, the Santa Fe Foundation, the Breast Cancer Research Foundation, and the ME Foundation. None of the funding sources influenced the design, interpretation or submission of this manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.plabm.2018.e00108.
Appendix A. Supplementary material
Supplementary material
References
- 1.Holley T., Lenkiewicz E., Evers L., Tembe W., Ruiz C., Gsponer J.R. Deep clonal profiling of formalin fixed paraffin embedded clinical samples. PLoS One. 2012;7:e50586. doi: 10.1371/journal.pone.0050586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musella V., Callari M., Di Buduo E., Scuro M., Dugo M., Miodini P. Use of formalin-fixed paraffin-embedded samples for gene expression studies in breast cancer patients. PLoS One. 2015;10:e0123194. doi: 10.1371/journal.pone.0123194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cravero K., Park B.H. Circulating tumor DNA-the potential of liquid biopsies. Curr. Breast Cancer Rep. 2016;8:14–21. [Google Scholar]
- 4.Quandt D., Dieter Zucht H., Amann A., Wulf-Goldenberg A., Borrebaeck C., Cannarile M. Implementing liquid biopsies into clinical decision making for cancer immunotherapy. Oncotarget. 2017;8:48507–48520. doi: 10.18632/oncotarget.17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi X., Ma J., Guan Y., Chen R., Yang L., Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int. J. Cancer. 2017;140:2642–2647. doi: 10.1002/ijc.30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 7.Tie J., Kinde I., Wang Y., Wong H.L., Roebert J., Christie M. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015;26:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi L., Pham T.H., Payabyab E.C., Sherry R.M., Rosenberg S.A., Raffeld M. Circulating tumor DNA as an early indicator of response to T-cell transfer immunotherapy in metastatic melanoma. Clin. Cancer Res. 2016;22:5480–5486. doi: 10.1158/1078-0432.CCR-16-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu D., Paoletti C., Gersch C., VanDenBerg D.A., Zabransky D.J., Cochran R.L. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin. Cancer Res. 2016;22:993–999. doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan K.C., Jiang P., Zheng Y.W., Liao G.J., Sun H., Wong J. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 11.Beaver J.A., Jelovac D., Balukrishna S., Cochran R.L., Croessmann S., Zabransky D.J. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res. 2014;20:2643–2650. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 13.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy S.R., Schmitt M.W., Fox E.J., Kohrn B.F., Salk J.J., Ahn E.H. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 2014;9:2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredebohm J., Mehnert D.H., Lober A.K., Holtrup F., van Rahden V., Angenendt P. Detection and quantification of KIT mutations in ctDNA by plasma safe-seqS. Adv. Exp. Med. Biol. 2016;924:187–189. doi: 10.1007/978-3-319-42044-8_34. [DOI] [PubMed] [Google Scholar]
- 16.Mohseni M., Cidado J., Croessmann S., Cravero K., Cimino-Mathews A., Wong H.Y. MACROD2 overexpression mediates estrogen independent growth and tamoxifen resistance in breast cancers. Proc. Natl. Acad. Sci. USA. 2014;111:17606–17611. doi: 10.1073/pnas.1408650111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19."Picard." Picard Tools - by Broad Institute: 〈http://picard.sourceforge.net〉.
- 20.Sherry S.T., Ward M., Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 21.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomczak K., Czerwinska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotoula V., Lyberopoulou A., Papadopoulou K., Charalambous E., Alexopoulou Z., Gakou C. Evaluation of two highly-multiplexed custom panels for massively parallel semiconductor sequencing on paraffin DNA. PLoS One. 2015;10:e0128818. doi: 10.1371/journal.pone.0128818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann U., Glockner S., Kleeberger W., von Wasielewski H.F., Kreipe H. Detection of gene amplification in archival breast cancer specimens by laser-assisted microdissection and quantitative real-time polymerase chain reaction. Am. J. Pathol. 2000;156:1855–1864. doi: 10.1016/S0002-9440(10)65059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacher A.G., Paweletz C., Dahlberg S.E., Alden R.S., O'Connell A., Feeney N. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2:1014–1022. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein A., Toro P.V., Lee J., Silberstein J.L., Nakazawa M., Waters I. Detection fidelity of AR mutations in plasma derived cell-free DNA. Oncotarget. 2017;8:15651–15662. doi: 10.18632/oncotarget.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christenson E.S., Dalton W.B., Chu D., Waters I., Cravero K., Zabransky D.J. Single-nucleotide polymorphism leading to false allelic fraction by droplet digital PCR. Clin. Chem. 2017;63:1370–1376. doi: 10.1373/clinchem.2017.273177. [DOI] [PubMed] [Google Scholar]
- 35.Orton R.J., Wright C.F., Morelli M.J., King D.J., Paton D.J., King D.P. Distinguishing low frequency mutations from RT-PCR and sequence errors in viral deep sequencing data. BMC Genom. 2015;16:229. doi: 10.1186/s12864-015-1456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinrich V., Stange J., Dickhaus T., Imkeller P., Kruger U., Bauer S. The allele distribution in next-generation sequencing data sets is accurately described as the result of a stochastic branching process. Nucleic Acids Res. 2012;40:2426–2431. doi: 10.1093/nar/gkr1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material