Figure 1.
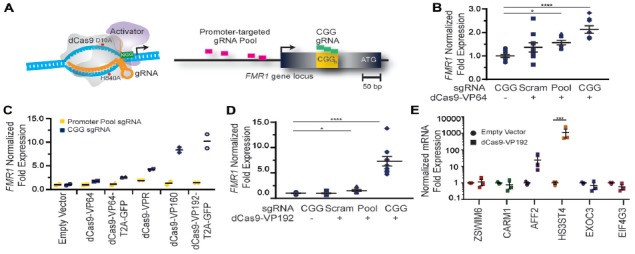
FMR1 messenger RNA (mRNA) increases with Clustered Regularly Interspaced Palindromic Repeat (CRISPR)-mediated targeting of transcriptional activators to either the FMR1 promoter or the CGG repeat. (A) Illustration of nuclease-inactive Cas9 (deficient Cas9, dCas9) fused to a transcriptional activator (left) and guide RNA (gRNAs) targeting regions within the FMR1 promoter or the CGG repeat (right). The promoter-targeted gRNA pool (pink) consisted of four gRNAs with unique targeting sequences, while the CGG gRNA (green) represents a single targeting sequence capable of tiling across the CGG repeat. (B) Relative FMR1 mRNA expression from three independent experiments in HEK293T cells transfected with empty vector or dCas9 fused to VP64 (dCas9-VP64) and non-targeting guide RNA (Scram), a pool of four guide RNAs within the FMR1 promoter (Pool), or a single CGG repeat guide (CGG). (C) Relative FMR1 mRNA expression from a single experiment in HEK293T cells transfected with an empty vector or dCas9 “second generation” activators and the FMR1 promoter gRNAs or the CGG gRNA. (D) Relative FMR1 mRNA expression from three independent experiments in HEK293T cells transfected with a control plasmid or dCas9 fused to VP192 (dCas9-VP192) and the indicated gRNA (for panels B–D, *p < 0.05, ****p < 0.0001, Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison test). (E) Relative expression of select CGG repeat-containing genes after transfection of HEK293T cells with CGG gRNA and dCas9-VP192 constructs (***p < 0.001, two-way ANOVA with Sidak’s multiple comparisons test). For all scatter plots shown, each data point represents an individual well and error bars on all graphs represent SEM.
