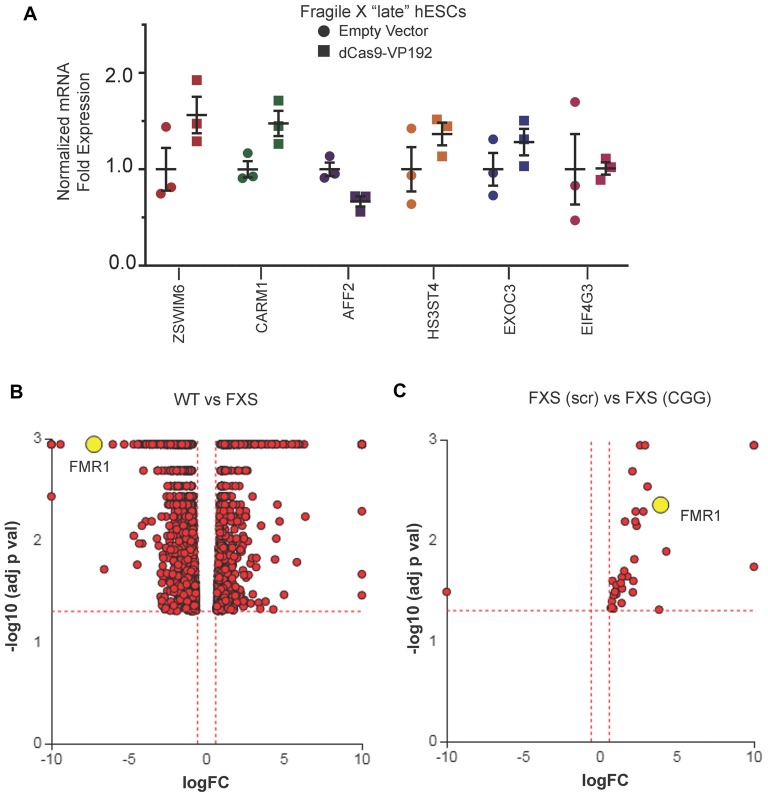
Figure 7.
Targeted reactivation of FMR1 by CGG guide RNAs has minimal off target effects compared to scramble guide RNA. (A) Relative fold expression of select CGG repeat-containing genes after transfection of late passage FXS hESCs with CGG gRNA and eGFP (empty vector) or dCas9-VP192 constructs. Each data point represents a technical replicate and error bars represent SEM. (B) Volcano plot showing RNA-seq analysis of WT and FXS hESCs. All 1784 significant differentially expressed (DE) genes are represented in terms of their measured fold change (x-axis) and the significance represented as the negative log (base 10) of the p-value (y-axis). The yellow dot shows the position of FMR1 mRNA. (C) Volcano plot showing RNA-seq analysis of FXS (scramble gRNA treated) and FXS (CGG gRNA treated) late passage hESCs expressing dCas9VP192. All 35 DE genes are represented as in terms of fold change. The axes are as described in (B). Yellow dot represents position of FMR1 mRNA. Images (B,C) were obtained from iPathwayGuide (http://www.advaitabio.com/ipathwayguide).